

Frank Morales is a Boeing Associate Technical Fellow /Technical Lead for Cloud-Interoperability Native Services at Boeing Global Services, Digital Solutions, and Analytics.
Thinkers360 Top Voices 2025
#1 Thought Leader: Open Source
#5 Thought Leader: Predictive Analytics
#6 Thought Leader: Agentic AI
#8 Thought Leader: Generative AI
#23 Thought Leader: Cryptocurrency
Top 100 Thought Leader: Agile, Artificial Intelligence, Healthcare, IT Strategy
In 1989, he received both B. Eng. and M. Eng. degrees in computer engineering, Avionics, and Artificial Intelligence with distinction from the Institute of Civil Aviation Engineers in Kyiv, Ukraine. He then became a 2001 senior member of IEEE. https://news.ieee.ca/2002/jan2002.htm#smupdates
Frank is a devout inventor, author, and speaker. He holds three US patents (7,092,748, 10,467,910, 10,522,045). He has published several technical peer-reviewed papers in prestigious journals such as Nature and authored a book chapter. He was a speaker at the 59th AGIFORS Annual Symposium with the theme entitled "Multi-Agent Systemic Approach to Support Dynamic Airline Operations based on Cloud Computing." His Google Scholar is here: https://scholar.google.com/citations?user=IlTdC5IAAAAJ&hl=en
He received several individual awards for his accomplishments with The Boeing Co. He also earned accreditation from the Massachusetts Institute of Technology (MIT) in the Sloan Executive Program Field of Study: Technology Strategies and Leadership.
He is a highly commended, analytical, and seasoned professional with a broad background in software and systems architecture, system integration, and project management. He possesses hands-on experience in business solutions architecture in the biomedical technology and aerospace industries. Demonstrate top-notch organizational skills in optimizing strategies to bridge the technical and business worlds while integrating technical solutions toward business problem resolutions.
I love the open-source community, and my GitHub repository for Machine/Deep Learning and AI is here:
https://github.com/frank-morales2020/MLxDL
He speaks fluent Spanish, Russian, and English.
Available For: Advising, Authoring, Consulting, Influencing, Speaking
Travels From: Montreal, Canada
Speaking Topics: Predictive Analytics & Machine Learning, Cloud Computing & Open Source, Generative AI
| FRANK MORALES | Points |
|---|---|
| Academic | 20 |
| Author | 676 |
| Influencer | 94 |
| Speaker | 3 |
| Entrepreneur | 150 |
| Total | 943 |
Points based upon Thinkers360 patent-pending algorithm.
 The Year of the Agent: A Retrospective on 2025’s AI Revolution
The Year of the Agent: A Retrospective on 2025’s AI Revolution
Tags: Agentic AI, Generative AI, Predictive Analytics
 Architecting Tomorrow's AI: A GPT-5.2 Multimodal API Sandbox
Architecting Tomorrow's AI: A GPT-5.2 Multimodal API Sandbox
Tags: Agentic AI, AGI, Generative AI
 The Architecture of Trust: How Gemini’s Deliberation Defines the Deep Research Agent
The Architecture of Trust: How Gemini’s Deliberation Defines the Deep Research Agent
Tags: Agentic AI, AGI, Generative AI
Tags: Agentic AI, Generative AI, Predictive Analytics
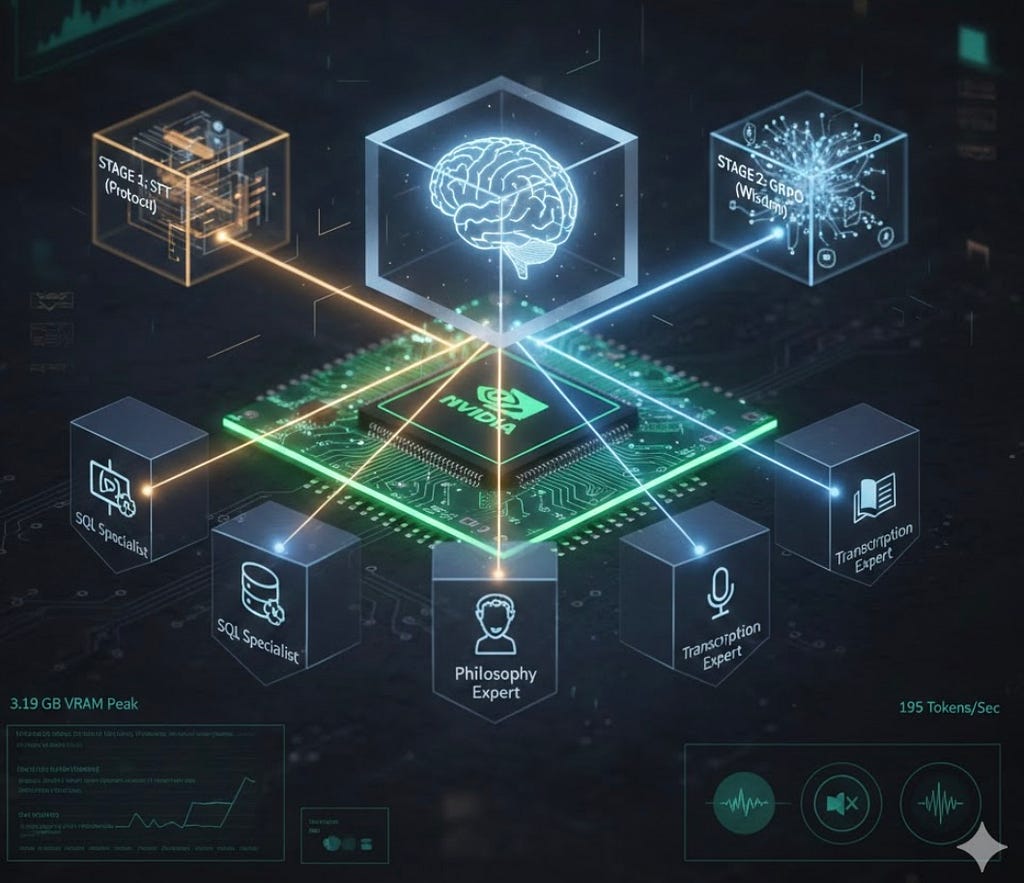 Intelligence Through Organization: Two-Stage Fine-Tuning for a High-Efficiency AI Orchestrator on…
Intelligence Through Organization: Two-Stage Fine-Tuning for a High-Efficiency AI Orchestrator on…
Tags: Agentic AI, Generative AI, Open Source
 The H2E Framework: Engineering Industrial Accountability into the Mistral-7B Era
The H2E Framework: Engineering Industrial Accountability into the Mistral-7B Era
Tags: Agentic AI, Generative AI, Open Source
 The H2E Framework: Engineering Industrial Accountability into the Mistral-7B Text-to-SQL Era
The H2E Framework: Engineering Industrial Accountability into the Mistral-7B Text-to-SQL Era
Tags: Agentic AI, Generative AI, Open Source
 The Evolution of Document Processing: The Recursive Language Model Framework
The Evolution of Document Processing: The Recursive Language Model Framework
Tags: Agentic AI, Generative AI, Open Source
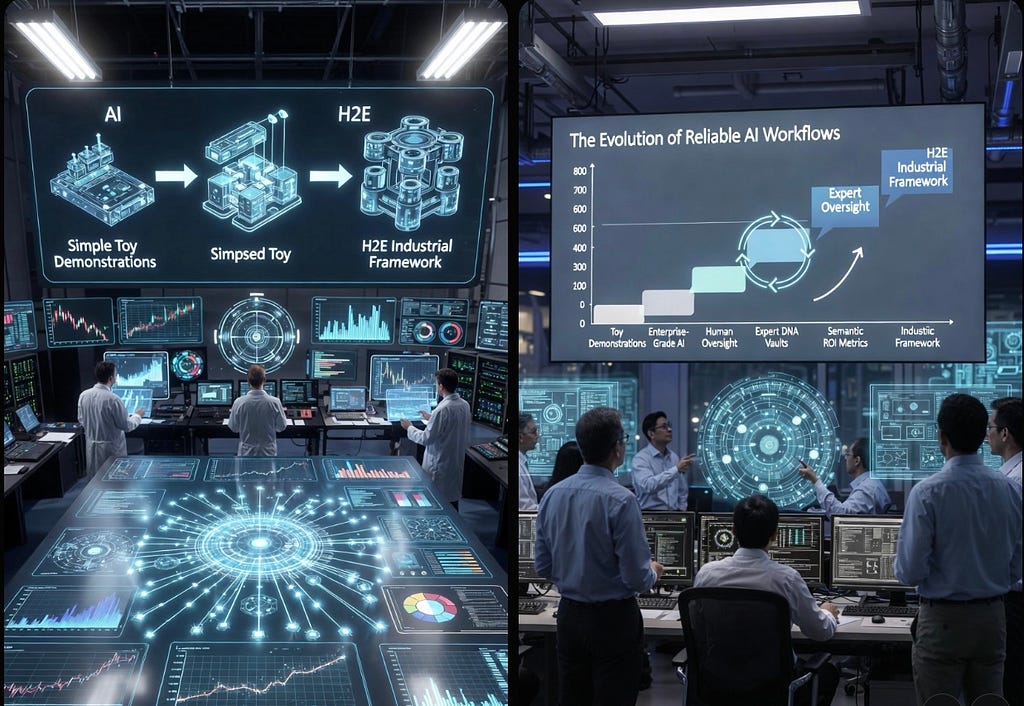 The Evolution of Reliable AI Workflows: From Toy Demonstrations to the H2E Industrial Framework
The Evolution of Reliable AI Workflows: From Toy Demonstrations to the H2E Industrial Framework
Tags: Agentic AI, Generative AI, Open Source
 The Open-Source Frontier: Control and Economic Sovereignty
The Open-Source Frontier: Control and Economic Sovereignty
Tags: Agentic AI, Generative AI, Open Source
 H2E: Engineering Provable Agency
H2E: Engineering Provable Agency
Tags: Agentic AI, Generative AI, Open Source
 The Architecture of Provable Agency: From Functional Autonomy to H2E Governance
The Architecture of Provable Agency: From Functional Autonomy to H2E Governance
Tags: Agentic AI, Generative AI, Open Source
 The Dawn of Medical AGI: Engineering Accountability through the H2E Framework
The Dawn of Medical AGI: Engineering Accountability through the H2E Framework
Tags: Agentic AI, Generative AI, Open Source
 Mistral and the Engineering of Provable Agency: The Convergence of Sovereign AI and the H2E…
Mistral and the Engineering of Provable Agency: The Convergence of Sovereign AI and the H2E…
Tags: Agentic AI, Generative AI, Open Source
 Engineering Provable Agency: The H2E Framework as a Deterministic Sentinel
Engineering Provable Agency: The H2E Framework as a Deterministic Sentinel
Tags: Agentic AI, Generative AI, Open Source
 The Sovereign Navigator: Implementing H2E Governance in Tesla’s FSD World Model
The Sovereign Navigator: Implementing H2E Governance in Tesla’s FSD World Model
Tags: Agentic AI, Generative AI, Open Source
 The H2E Framework in Action: Engineering Accountability Through Code with Mistral-7B
The H2E Framework in Action: Engineering Accountability Through Code with Mistral-7B
Tags: Agentic AI, Generative AI, Open Source
 The Sovereign Driver: How the Waymo World Model Redefines Autonomy
The Sovereign Driver: How the Waymo World Model Redefines Autonomy
Tags: Agentic AI, Generative AI, Open Source
 The Sovereign Shield: Mitigating Model Collapse and Diversity Decay through Strategic Autonomy
The Sovereign Shield: Mitigating Model Collapse and Diversity Decay through Strategic Autonomy
Tags: Agentic AI, Generative AI, Open Source
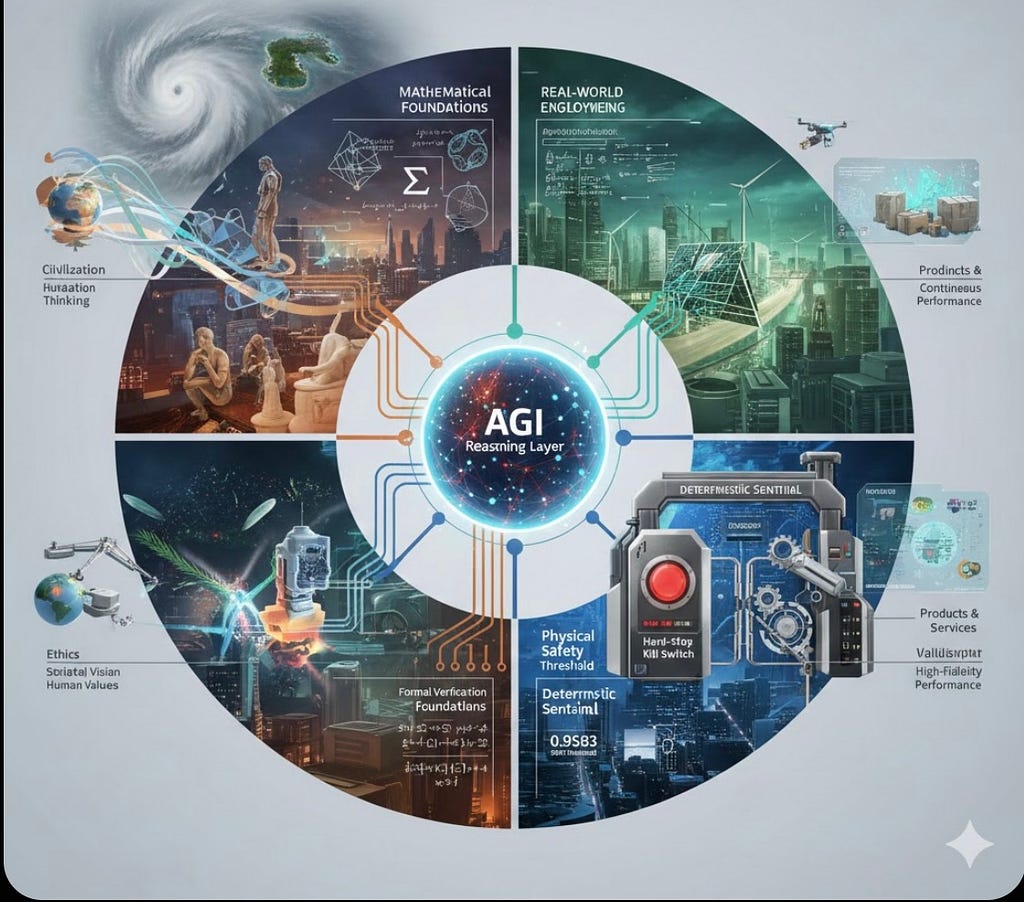 The H2E Industrial Ecosystem: Engineering Accountable Agency for Global Crises
The H2E Industrial Ecosystem: Engineering Accountable Agency for Global Crises
Tags: Agentic AI, Generative AI, Open Source
 DNA of Flight: Human-to-Expert (H2E) Governance for Autonomous Skies
DNA of Flight: Human-to-Expert (H2E) Governance for Autonomous Skies
Tags: Agentic AI, Generative AI, Open Source
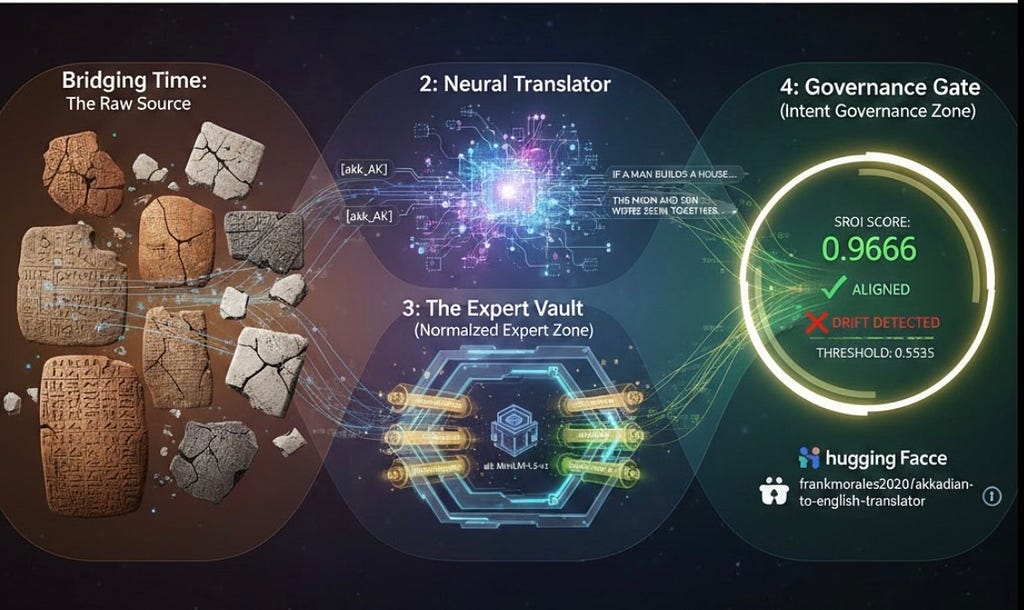 Bridging 4,500 Years: How H2E Turned an Ancient Language into a Verifiable, Sovereign AI Translator
Bridging 4,500 Years: How H2E Turned an Ancient Language into a Verifiable, Sovereign AI Translator
Tags: Agentic AI, Generative AI, Open Source
 NeMo-Driven Sovereignty: Precision Fine-Tuning and Algorithmic Governance in Llama-3
NeMo-Driven Sovereignty: Precision Fine-Tuning and Algorithmic Governance in Llama-3
Tags: Agentic AI, Generative AI, Open Source
 Claude 4.6 + H2E: Building a Governed Multi-Agent System with 86% Alignment at $14.80
Claude 4.6 + H2E: Building a Governed Multi-Agent System with 86% Alignment at $14.80
Tags: Agentic AI, Generative AI, Open Source
 Engineering Accountability: Constructing Deterministic AI in a Probabilistic World
Engineering Accountability: Constructing Deterministic AI in a Probabilistic World
Tags: Agentic AI, Generative AI, Open Source
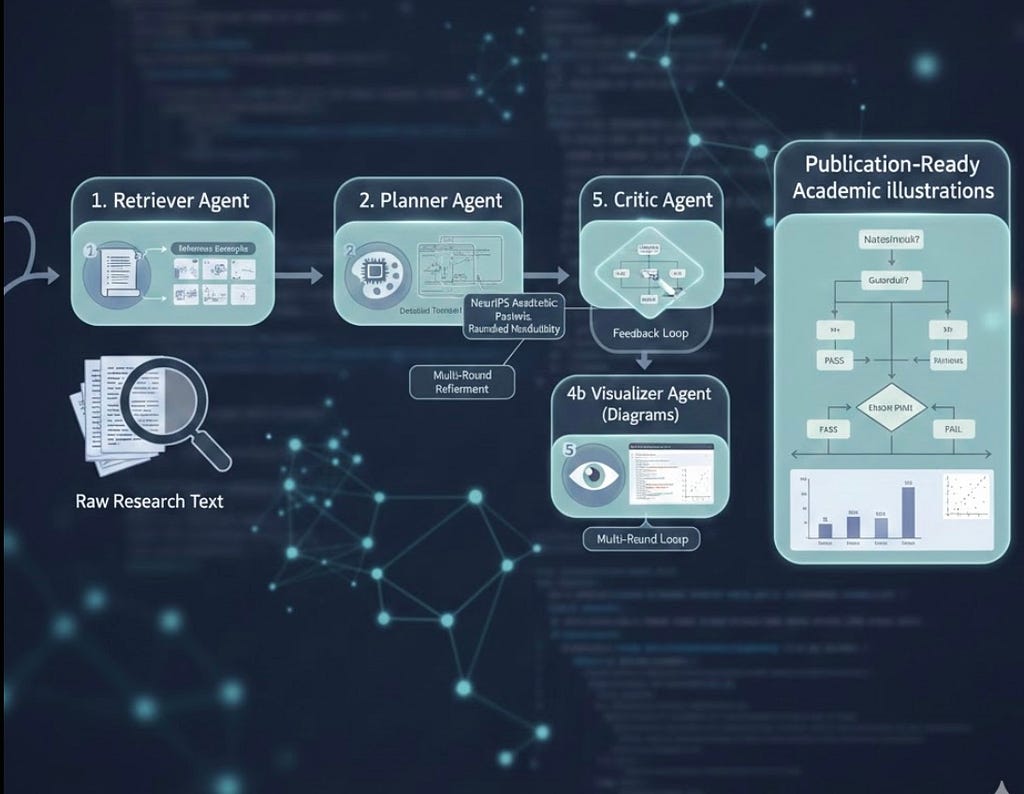 The Evolution of Autonomous Research Communication: An Analysis of PaperBanana
The Evolution of Autonomous Research Communication: An Analysis of PaperBanana
Tags: Agentic AI, Generative AI, Open Source
 The Rise of Sovereign AI: Engineering Determinism in a Probabilistic World
The Rise of Sovereign AI: Engineering Determinism in a Probabilistic World
Tags: Agentic AI, Generative AI, Open Source
 The Architecture of Accountability: A NeMo-Based Text-to-SQL POC
The Architecture of Accountability: A NeMo-Based Text-to-SQL POC
Tags: Agentic AI, Generative AI, Open Source
 The Dawn of Agentic Finance: Governance through the H2E Framework
The Dawn of Agentic Finance: Governance through the H2E Framework
Tags: Agentic AI, Generative AI, Open Source
Tags: Agentic AI, AI, Generative AI
Tags: Agentic AI, AI, Generative AI
 Program Certificate - Executive Certificate in Management and Leadership
Program Certificate - Executive Certificate in Management and Leadership
Credential ID https://www.linkedin.com/in/frank-morales1964/overlay/1635475339334/single-media-viewer/?profileId=A
Tags: Agentic AI, AI, Open Source
Tags: Agentic AI, AI, Generative AI
Tags: AI, Analytics, Predictive Analytics
Tags: Agile, Analytics, Generative AI
Tags: AI, Analytics, Predictive Analytics
Tags: AI, Generative AI, Predictive Analytics
Patent Number 10467910 and United States Patent 10522045
Tags: Agentic AI, Generative AI, Predictive Analytics
Patent Number United States Patent 7092748
Tags: Agentic AI, Generative AI, Predictive Analytics
Tags: Agentic AI, Open Source, Predictive Analytics
Tags: Healthcare, Predictive Analytics
Tags: Healthcare
Tags: Healthcare
 Multi-Agent Systemic Approach to Support Dynamic Airline Operations based on Cloud Computing
Multi-Agent Systemic Approach to Support Dynamic Airline Operations based on Cloud Computing
Tags: Agentic AI, AI, Predictive Analytics
Date : November 03, 2025
Date : November 03, 2025
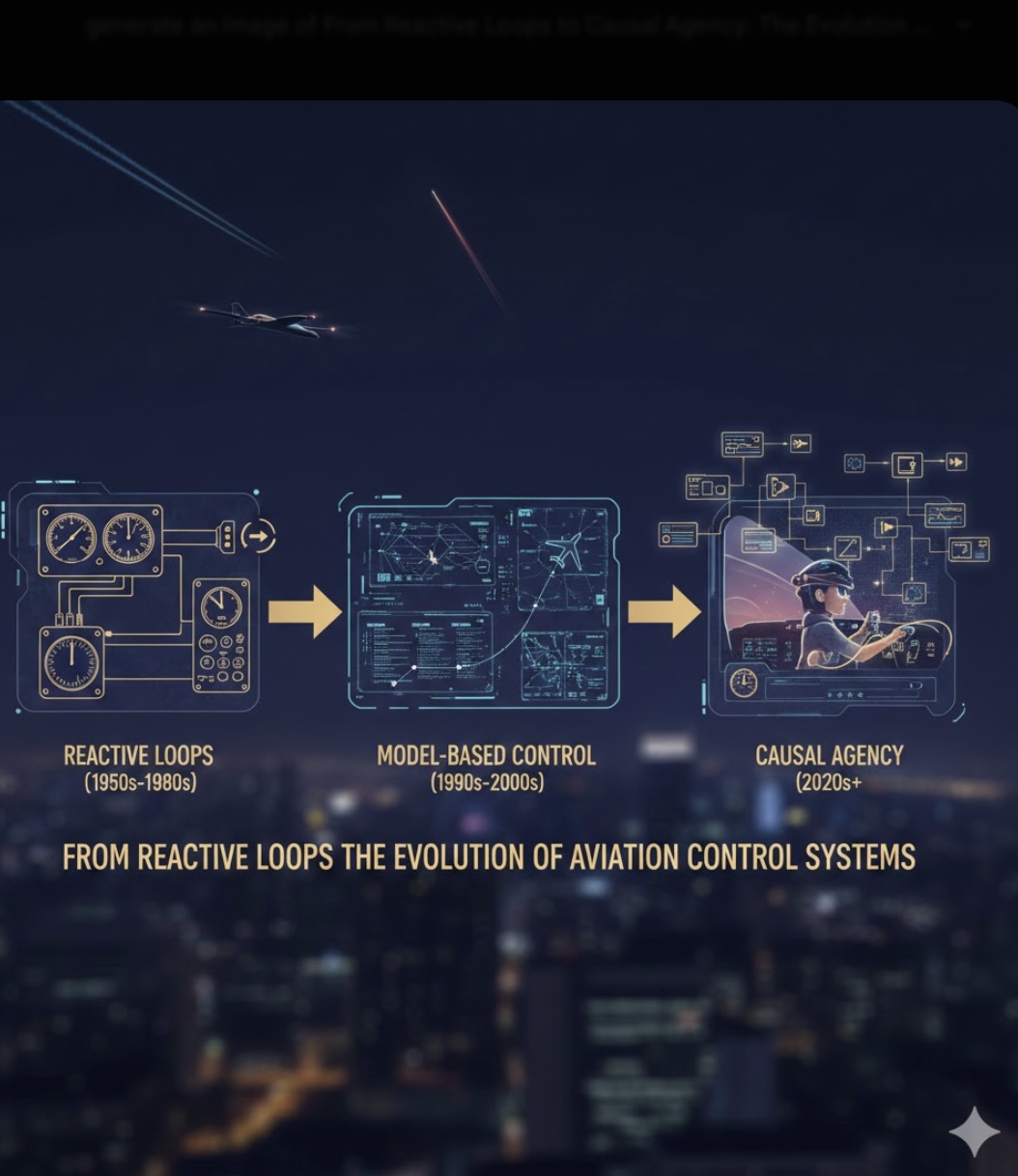 From Reactive Loops to Causal Agency: The Evolution of Aviation Control Systems
From Reactive Loops to Causal Agency: The Evolution of Aviation Control Systems
The transition from classical aviation control to the architecture presented in the LEJEPA_VJEPA_AGI_DEMO.ipynb notebook represents a fundamental shift from reactive error-correction to proactive, world-model-based reasoning. While traditional systems focus on correcting immediate errors, this architecture focuses on predicting future physical states and understanding the causal "why" behind flight events.
LEJEPA_VJEPA_AGI_DEMO.ipynb: https://github.com/frank-morales2020/MLxDL/blob/main/LEJEPA_VJEPA_AGI_DEMO.ipynb
| Feature | Traditional PID / Autopilot | Causal Planning (JEPA-based) |
| Core Logic | Reactive: Calculates a "tracking error" and applies gains to minimize it. | Proactive: Simulates future states in a latent "world model" to select the best action sequence. |
| Knowledge | Implicit: Operates on mathematical derivatives without "knowing" flight concepts. | Explicit: Uses a "modular hybrid cognitive stack" to ground physics in semantic concepts. |
| Data Handling | Point-in-Time: Processes immediate sensor input (altitude, speed) to adjust surfaces. | Spatio-Temporal: Analyzes video sequences and historical trajectories to understand dynamics. |
| Failure Mode | Disengagement: Often defaults to "disengage and alert" when sensor data is conflicting. | Graceful Reasoning: Uses an LLM to provide a causal assessment of anomalies and suggest fixes. |
1. From Correlation to Causality
Traditional autopilots are constrained by the frequency-domain tuning of their PID loops, which respond primarily to events, such as a drop in altitude. The Morales framework uses the DeepSeek-reasoner to interpret why an event occurs—for example, identifying engine power loss during a final approach—bridging the gap between raw telemetry and symbolic causal inference.
2. Eliminating Control "Hacks" with SIGReg
Traditional robust control requires complex mathematical development and manual tuning. The implementation of SIGReg (Sketched Isotropic Gaussian Regularization) simplifies this process by enforcing stable $N(0,I)$ latent distributions without the need for momentum teachers or stop-gradients. This mechanism effectively prevents "representational collapse," a common failure mode in earlier AI-driven controllers.
3. Model Predictive Path Planning (MPPI)
Unlike a PID controller that acts on a single setpoint, the Predictive Latent Dynamics Model (PLDM) allows for "System II" cognitive processing. This involves running a "simulation-in-the-head" to project 4D aircraft states into the future. By evaluating multiple "what-if" scenarios before the actual control surfaces move, the agent mimics the high-level planning a human pilot performs during emergency procedures.
The notebook addresses representational hurdles by adopting the November 2025 LeJEPA framework. SIGReg enforces stable statistics on latent representations, eliminating the complex heuristics used in earlier self-supervised models. Furthermore, by leveraging a frozen V-JEPA backbone for feature extraction and DeepSeek for semantic assessment, the architecture provides a natural-language causal analysis of flight phases.
The shift toward AI-driven engineering agency marks an era where flight systems possess a "Physical DNA" of their environment. By combining the visual perception of V-JEPA with the stabilized physical forecasting of LeJEPA, this architecture moves aviation closer to truly autonomous agents that understand the causal physics governing safety-critical domains.
Tags: Predictive Analytics, Generative AI, Agentic AI
 The Digital Navigator: The Role of Artificial Intelligence in Artemis II
The Digital Navigator: The Role of Artificial Intelligence in Artemis II
As the Artemis II mission prepares to carry humanity back to the vicinity of the Moon, it represents a fundamental shift in how we explore deep space. While the primary mission objective is to validate the safety and performance of the Space Launch System (SLS) and the Orion spacecraft for human travel, the invisible engine driving this validation is Artificial Intelligence. Unlike the rigid software of the Apollo era, Artemis II utilizes AI as a dy"amic "fifth crew member, bridging the gap between human intuition and the overwhelming data density of modern spaceflight.
The Artemis II mission is currently in its final pre-launch phase at the Kennedy Space Center. As of today, January 19, 2026, the mission has reached a major milestone: the SLS rocket and Orion spacecraft were successfully rolled out to Launch Pad 39B this past weekend, arriving on January 17 after a nearly 12-hour journey from the Vehicle Assembly Building.
The mission is currently tracking toward the following timeline:
This mission carries a diverse crew of four who will be the first humans to travel to the vicinity of the Moon in over 50 years. The crew includes Commander Reid Wiseman, Pilot Victor Glover, and Mission Specialists Christina Koch and Jeremy Hansen. Glover will be the first person of colour, Koch the first woman, and Hansen the first non-American to fly a lunar mission.
Artemis II is a crewed flyby, meaning the astronauts will not land on the Moon. Instead, they will:
The Artemis II mission is supported by a massive industrial base, involving over 3,800 suppliers across all 50 U.S. states and several international partners. While NASA leads the mission, the hardware and ground systems are built and managed by several prime aerospace contractors.
Core Mission Partners
Key Infrastructure and Technology Providers
Beyond the main rocket and capsule, several other companies provide critical mission support. L3Harris provides the mission-critical audio system and various avionics systems. United Launch Alliance provided the upper stage used to propel Orion toward the Moon. MDA Space, a major Canadian partner, provides technical support and is the lead for future lunar robotics. Companies like Bechtel and Jacobs provide the engineering for mobile launchers and ground system support.
Deep space navigation presents a unique challenge: once Orion leaves Earth’s orbit, traditional GPS becomes unavailable. To maintain a precise trajectory, the spacecraft relies on AI-driven Optical Navigation.
This system utilizes high-resolution cameras to capture images of the Moon and Earth against the backdrop of stars. AI algorithms process these data points in real time, identifying celestial bodies and cross-referencing them with preloaded star maps. This allows the spacecraft to determine its position and velocity autonomously, independent of ground control. Furthermore, during proximity operations, AI provides the necessary stabilization logic, ensuring that human steering inputs are executed with precision.
The Orion spacecraft is equipped with hundreds of thousands of sensors monitoring everything from cabin pressure to electrical health. AI-driven anomaly detection systems move beyond simple threshold-based alerts by analyzing nonlinear relationships across multiple sensors. If a slight increase in power draw correlates with a minor temperature shift, the AI can flag a component for degradation well before a failure. This proactive approach to health management allows the team to address issues during quiet flight phases rather than during high-stakes maneuvers.
AI also plays a critical role in managing the health and performance of the astronauts. Using wearable devices, AI analyzes crew members' sleep patterns, stress levels, and cognitive performance to help mission control optimize flight schedules. Additionally, NASA is testing intelligent interfaces that allow the crew to access technical manuals and spacecraft status reports using natural language, significantly reducing their cognitive load.
This mission serves as the ultimate stress test for the hardware and procedures that will be used for Artemis III, which is currently planned to land the first woman and first person of colour on the lunar surface as early as 2027. By integrating AI into its fabric, NASA is ensuring that, as humans travel further into the cosmos, they are supported by a digital infrastructure as resilient and adaptable as the explorers themselves.
This video provides an excellent visual overview of the Artemis II mission timeline and the roles of the various crew members and partner organizations.
Tags: Agentic AI, Generative AI, Predictive Analytics
 The Wireless Renaissance: From Tesla’s Dream to Agentic Autonomy
The Wireless Renaissance: From Tesla’s Dream to Agentic Autonomy
For over a century, the concept of wireless power transmission resided in the realm of visionary speculation and laboratory curiosity. Nikola Tesla, the father of the modern electrical age, famously dreamed of a "World Wireless System" where the Earth and its atmosphere would act as conductors, delivering energy to any point on the globe without a single foot of copper wire.1 Today, that dream is being realized not as a single global monolith, but as a sophisticated suite of technologies—lasers, ultrasonics, and radio-frequency harvesting—that are poised to untether our most advanced intelligence: Agentic AI.
The recent breakthroughs from researchers at the University of Helsinki and the University of Oulu represent a paradigm shift in how we power autonomous systems. By using high-intensity ultrasonic sound waves to create "acoustic wires"—channels of low-density air that guide electrical sparks—science has found a way to "beam" physical electricity.
For Agentic AI, this is the missing piece of the physical-layer puzzle. Until now, the "autonomy" of an AI agent was strictly limited by its battery capacity (the "Battery Tax"). In complex Multi-Agent Systems (MAS), such as a swarm of drones or a robotic banking security team, the need to return to a charging dock creates a massive operational gap. Wireless power transfer (WPT) allows these agents to move from "rechargeable" to "perpetual."
The impact on aerospace and formation flight is particularly profound. In a multi-agent aerial environment, traditional refuelling or recharging is a dangerous and complex maneuver. Wireless power changes the fundamental physics of the mission:
Formation-Based Recharging: A lead aircraft, acting as a "power hub," could use laser-based "power-by-light" systems to transmit energy to smaller trailing agents. This ensures that the formation can remain aloft indefinitely, optimized by AI to minimize drag and maximize energy reception.
Galvanic Isolation in High-Voltage Zones: In aerospace testing and nuclear environments, physical wires are a liability. Wireless energy provides a "firewall for physics," allowing AI monitoring agents to operate in high-radiation or high-voltage zones without the risk of a surge traveling back through a cable to fry the central processing unit.
If wireless power gives AI freedom, AI gives wireless power efficiency. The greatest challenge of WPT has always been alignment; even a slight movement can cause the energy beam to miss its mark.
Modern Agentic AI serves as the real-time "pilot" for these energy beams. Using machine learning-driven beamforming, the AI can predict the trajectory of a moving drone or robot and micro-adjust the ultrasonic or laser emitter in milliseconds. This transforms a "dumb" broadcast into a high-precision, goal-oriented delivery system.
In 1926, Tesla predicted a world where a man could carry a device in his pocket, powered and connected wirelessly, capable of seeing and hearing across the world. While we have achieved the "connected" part through Wi-Fi and 5G, we are only now achieving the "powered" part.
The transition to a cable-free infrastructure is more than a convenience; it is the birth of perpetual autonomy. By combining the raw power of Finnish "acoustic wires" with the cognitive reasoning of Agentic AI, we are finally building the world Tesla saw: a world where energy is as ambient and accessible as the air we breathe.
The Secret of Nikola Tesla's Wireless Power
This video explores the practical engineering behind laser-based power beaming and how it is being used to keep drones in the air for kilometres at a time, bringing Tesla's theories into the 21st-century sky.
Tags: Agentic AI, Generative AI, Predictive Analytics
 Autonomous Wingmen: Scaling Sustainable Aviation via NVIDIA NAT and Formation Flight
Autonomous Wingmen: Scaling Sustainable Aviation via NVIDIA NAT and Formation Flight
The aviation industry stands at a critical juncture, facing the dual challenge of meeting rising global travel demand while drastically reducing its environmental footprint. Traditional efficiency gains, once driven primarily by jet engine evolution, are reaching a plateau, necessitating radical aerodynamic and operational innovations. One of the most promising solutions is aerodynamic formation flight—a biomimetic strategy inspired by migrating birds that allows trailing aircraft to "surf" the upwash of a lead aircraft's wingtip vortices2. By integrating this concept with Multi-Agent Systems (MAS) and Large Language Models (LLMs), the industry can move toward a highly optimized, automated, and sustainable transatlantic corridor.
At its core, formation flight is an energy-saving mechanism. When a follower aircraft positions itself precisely within the upwash generated by a leader, it leverages "wake energy retrieval" to reduce induced drag and the thrust required for cruise flight.
The operational execution of pairing two aircraft mid-flight presents a staggering coordination challenge. Traditional centralized automation often lacks the flexibility to manage the real-time variables of the North Atlantic Track (NAT) system.
The operational logic of formation flight is driven by a sophisticated Multi-Agent Systems framework, specifically using tools such as the NVIDIA NAT (NeMo Agent Toolkit). The system's architecture is built on a modular "Contract-First" design, where structured data models define the parameters for every automated decision.
1. Structured Data Modelling
The architecture's foundation lies in rigorous data validation with Pydantic. Primary models act as specialized contracts for the system's agents:
2. Specialized Multi-Agent Logic
The system employs distinct functions that operate as independent micro-agents:
3. Asynchronous Mission Orchestration
A central execution engine utilizes asynchronous programming to coordinate these agents:
The complete implementation of this multi-agent logic is available in the full code on GitHub: https://github.com/frank-morales2020/MLxDL/blob/main/NAT_FormationFlightPairing_DEMO.ipynb.
While automated systems handle technical orchestration, Large Language Models (LLMs) serve as the critical interface between these systems and human professionals. Advanced simulations generate NAT Formation Dispatch Reports that combine technical flight data with generative AI to produce professional briefing bulletins.
1. Flight Dispatch Bulletins
Generative models produce distinct reports based on mission results:
2. Fuel Analysis Results
Simulations provide a quantitative comparison of fuel consumption:
The operational concepts detailed in this architecture align with the latest sustainability milestones in the aviation industry. Global carriers are actively transitioning from theoretical research to live operational trials. For instance, recent progress reports highlight successful trans-Atlantic flight trials and the validation of pairing technologies that safely guide aircraft to precise rendezvous points. These advancements are a core part of broader decarbonization goals, which include investing in next-generation aircraft and scaling Sustainable Aviation Fuel (SAF)
Detailed insights into these real-world sustainability milestones can be found here: https://news.delta.com/ground-and-air-we-keep-climbing-deltas-year-sustainability-progress.
Conclusion: A New Standard for the Skies
The integration of aerodynamic formation flight with AI-driven orchestration represents more than just a technical achievement; it is a necessary evolution for a hard-to-decarbonize industry. By leveraging the natural energy-saving principles of migratory birds and the computational power of multi-agent intelligence, the aviation sector can realize substantial fuel savings and move closer to its 2050 goal of net-zero emissions. As these technologies mature, the North Atlantic will transform from a series of isolated solo tracks into a synchronized, efficient, and sustainable network.
Tags: Agentic AI, Generative AI, Predictive Analytics
 Building the Foundation for Agentic AI: A Demonstration of NVIDIA’s NeMo Agent Toolkit (NAT)
Building the Foundation for Agentic AI: A Demonstration of NVIDIA’s NeMo Agent Toolkit (NAT)
The emergence of Large Language Models (LLMs) has shifted the focus of AI development from simple chatbots to autonomous "agents"—systems capable of reasoning, planning, and executing complex tasks by interacting with external tools. At the forefront of this evolution is NVIDIA's NeMo Agent Toolkit (NAT), an open-source library for building, profiling, and optimizing high-performance AI agent workflows. The provided demonstration notebooks illustrate a critical "Day 1" workflow: preparing standalone Python tools and seamlessly integrating them into a managed agentic system.
NAT serves as a framework-agnostic "glue" layer, allowing developers to connect various LLMs with specialized functional tools. Unlike monolithic systems, NAT encourages a modular approach. As demonstrated in the notebooks, the first step in building a NAT agent is creating "Standalone Tools"—standard Python functions that remain independent of the toolkit until they are registered. In these examples, the tools are designed for climate analysis, capable of loading NOAA temperature records, calculating statistical trends, and generating visualizations like annual anomaly plots.
Using Google Colab as the primary environment highlights the toolkit's accessibility and integration with cloud workflows. The notebooks leverage colab_env to manage secure environment variables, specifically the NVIDIA_API_KEY, which provides access to NVIDIA NIMs (Inference Microservices). By programmatically creating a local module (climate_tools_simple.py) and updating the system path, the demonstration shows how a temporary cloud environment can be transformed into a robust development platform for AI agents.
The demonstration notebooks are designed to showcase the versatility and framework-agnostic nature of NAT. A key goal of these demos is to prove that the same open-source toolkit can seamlessly manage both commercial and open-source Large Language Models (LLMs) within a unified workflow.
Dual-Model Integration Strategy
The notebooks achieve this by utilizing the same backend "Tools" and infrastructure while swapping the "Reasoning Engine" (the LLM):
Commercial LLM Integration: The first notebook focuses on integrating a commercial LLM, specifically GPT-4, as the reasoning engine. This demonstrates how NAT can act as a secure bridge for high-performance, proprietary models.
Open-Source LLM Integration: The second notebook, DEEPSEEK_NAT_DEMO_JAN2025.ipynb, focuses on integrating DeepSeek, a prominent open-source model. It shows that the toolkit can successfully deploy open-source models to perform the same complex data analysis tasks as their commercial counterparts.
DEEPSEEK_NAT_DEMO_JAN2025.ipynb: https://github.com/frank-morales2020/MLxDL/blob/main/DEEPSEEK_NAT_DEMO_JAN2025.ipynb
/NEMO_Equation_AAI_DEMO.ipynb: https://github.com/frank-morales2020/Cloud_curious/blob/master/NEMO_Equation_AAI_DEMO.ipynb
Consistent Toolkit, Different Models
By using the NeMo Agent Toolkit as the constant factor, the demos illustrate several technical advantages:
Unified Configuration: Both models use a similar YAML-based configuration (config.yml) to define the agent's behaviour and the tools it can access.
Shared Tooling: Both the GPT-4 and DeepSeek agents leverage the same standalone Python module (climate_tools_simple.py) for climate data loading, statistical analysis, and visualization.
Environment Management: Both demos utilize colab_env and NVIDIA_API_KEY to securely manage model access, whether connecting to NVIDIA-hosted open-source NIMs or commercial endpoints.
This approach emphasizes that NAT is a glue layer that allows developers to choose the best model for their specific needs—whether open-source for transparency or commercial for performance—without rebuilding their entire agentic infrastructure.
The true power of NAT is realized when these local Python functions are bridged with an LLM's reasoning capabilities. In the DeepSeek iteration of the demo, the agent follows a structured process to answer natural language queries like "Find the warmest year between 1980 and 2000":
Reasoning: It identifies the need for statistical analysis.
Tool Execution: It calls the find_extreme_years function from the standalone module.
Synthesizing: It processes the tool output to provide a clear, factual answer, such as identifying 1998 as the warmest year with a 0.79°C anomaly.
The NAT demonstration notebooks provide a blueprint for modern AI development. By separating the "brain" (the LLM) from the "hands" (the Python tools), and using NAT to orchestrate their interaction, developers can create reliable, verifiable, and highly specialized agents. Whether analyzing global climate trends or managing complex industrial data, NVIDIA's NeMo Agent Toolkit offers the necessary infrastructure to move AI from experimental code to impactful, real-world applications.
Tags: Predictive Analytics, Generative AI, Agentic AI
 The Architect of Agency: NVIDIA’s Vera CPU and the Dawn of the AI Super-Factory
The Architect of Agency: NVIDIA’s Vera CPU and the Dawn of the AI Super-Factory
In the rapidly evolving landscape of artificial intelligence, the transition from "chatbots" to "autonomous agents" has necessitated a fundamental rethinking of computer architecture. At CES 2026, NVIDIA signalled the end of the general-purpose era in data centers with the unveiling of the Vera CPU. More than just a processor, Vera is a custom-engineered "data engine" designed to eliminate the bottlenecks that have long prevented AI from achieving actual, real-time reasoning at scale. By moving from off-the-shelf components to the custom "Olympus" core, NVIDIA has not only doubled performance but has redefined the role of the CPU in the modern AI factory.
The defining characteristic of the Vera CPU is the Olympus core, NVIDIA's first fully bespoke implementation of the Armv9.2-A instruction set. While its predecessor, Grace, relied on standard Arm Neoverse designs, Olympus is a ground-up reimagining of what a CPU core should do in an AI-centric world.
The core's efficiency stems from its expanded math capabilities. Each of the 88 Olympus cores features six 128-bit SVE2 vector engines, a 50% increase over Grace. More importantly, it is the first CPU to support FP8 precision natively. By processing data in the same 8-bit format used by the latest GPUs, Vera can move and manipulate AI data without the "translation tax" of converting between different formats, drastically reducing latency during the critical pre-fill stages of model inference.
While the hardware specifications of the Vera CPU are formidable, its impact is felt at the software layer—specifically through native support for FP8 (8-bit floating-point) precision. Historically, CPUs have operated in high-precision formats such as FP32 and FP64. While accurate, these formats are computationally "heavy" and memory-intensive. In contrast, AI training and inference have increasingly shifted toward lower precision to achieve greater speed. By bringing FP8 support to the Olympus core, NVIDIA has effectively taught the CPU and GPU to speak the same mathematical language.
In previous generations, a significant amount of "compute overhead" was wasted on data casting. When a CPU prepared data for a GPU, it often had to convert FP32 numbers down to FP8 or INT8. This conversion layer introduced latency and increased power consumption.
With Vera, the Olympus cores can process FP8 natively. This means that during the pre-fill stage of a Large Language Model—where the CPU parses input text and prepares the initial tensors—the data remains in its optimized AI format from the moment it hits the CPU until it reaches the GPU. This "lossless" transition in format results in a dramatic increase in system-wide efficiency.
For developers, the inclusion of FP8 on the CPU side fundamentally alters the CUDA development workflow. Traditionally, programmers had to manage "precision boundaries carefully"—deciding exactly where to downscale data to avoid losing accuracy while maintaining speed.
Unified Data Types: Developers can now define a single FP8 tensor that spans both CPU and GPU memory spaces. This simplifies the code significantly, as the cudaMemcpy Functions no longer require an intermediate conversion kernel.
Simplified Quantization: NVIDIA's Transformer Engine software can now manage quantization (the process of shrinking data) across the entire NVL72 rack. Because the Vera CPU supports FP8, the Transformer Engine can dynamically scale precision based on the "importance" of the data, keeping critical weights at higher precision while moving transient data to FP8.
Faster Debugging and Profiling: Since the CPU can now run FP8 kernels natively, developers can profile and debug AI logic on the CPU using the same data formats that will eventually run on the GPU. This reduces the "it works on CPU but fails on GPU" errors that have plagued AI engineering.
The switch to FP8 isn't just a software convenience; it radically changes the physics of data movement. On the Vera platform, the benefits of FP8 over traditional 16-bit and 32-bit formats are quantifiable:
| Precision Format | Bits per Value | Relative Memory Footprint | Bandwidth Efficiency | Accuracy Retention (LLMs) |
| FP32 (Single) | 32 bits | 4x | 25% (Baseline) | 100% (Gold Standard) |
| FP16 / BF16 | 16 bits | 2x | 50% | ~99.9% |
| FP8 (Vera Native) | 8 bits | 1x | 100% | ~99.5%* |
> Note: Accuracy retention for FP8 is maintained via NVIDIA's Transformer Engine, which uses dynamic scaling factors to prevent numerical underflow.
Perhaps the most technically provocative feature of the Vera CPU is Spatial Multi-Threading (SMT). Traditional multi-threading, which has dominated computing for decades, works by "time-slicing"—alternating between two tasks so quickly it creates the illusion of simultaneity. However, in high-stakes AI workloads, this can lead to "resource contention," where one thread stalls while waiting for the other to release the core's assets.
Vera's Spatial SMT takes a different approach by physically partitioning the core's internal execution ports. Rather than sharing the same hardware over time, the two threads occupy separate physical lanes within the core. This ensures "deterministic performance," allowing the system to handle 176 simultaneous threads with predictable latency.
The most significant bottleneck in modern Large Language Models (LLMs) is not math, but memory—specifically the KV-cache. As AI conversations grow longer or involve large documents, the "Key-Value" data that represents the model's short-term memory can expand until it overflows the GPU's expensive High Bandwidth Memory (HBM).
The Vera CPU addresses this with a massive 1.5 TB LPDDR5X memory pool, a 3x increase over the previous generation. Through the 1.8 TB/s NVLink-C2C interconnect, Vera functions as a "Context Memory Storage" tier. When a GPU's memory is full, it can offload the KV-cache to the Vera CPU at nearly 7x the speed of traditional PCIe connections. This allows AI agents to "remember" hundreds of pages of context without the performance hit of recomputing data from scratch.
By integrating FP8 into the very heart of the Olympus core, NVIDIA has removed the "translation tax" that has hindered heterogeneous computing for years. This alignment allows the Vera CPU to act as a true co-processor, handling complex logic and data preparation at the same velocity as the GPUs. The result is a software environment where the hardware becomes transparent, allowing developers to focus on the complexity of their AI agents rather than the minutiae of bit-depth management.
Tags: Agentic AI, Generative AI, Predictive Analytics
 The Resurgence of 1967 Mathematics: How DeepSeek Stabilized the AI of 2026
The Resurgence of 1967 Mathematics: How DeepSeek Stabilized the AI of 2026
In January 2026, DeepSeek researchers published a landmark paper titled "mHC: Manifold-Constrained Hyper-Connections," solving a "foundational instability" problem that had previously limited the depth and complexity of AI models. This breakthrough centers on the Sinkhorn-Knopp algorithm, a piece of linear algebra from 1967, which DeepSeek repurposed to ensure that signals remain numerically stable even in stacks hundreds of layers deep. By bridging nearly sixty years of mathematical theory with cutting-edge GPU engineering, DeepSeek has unlocked a pathway for the next generation of reasoning-first AI.
Since 2015, the industry standard for neural networks has been Residual Connections (ResNet), which provides a "highway" for information to skip through layers unchanged, preventing signals from fading. In late 2024, researchers introduced Hyper-Connections (HC)—a "multi-lane" version of this highway that allowed for richer mixing and more flexible information routing.
The Failure: While Hyper-Connections increased a model's expressive power, they were notoriously unstable. Without constraints, signal "energy" could be amplified by over 3,000x as it passed through deep networks. This frequently resulted in "loss spikes" and "NaN" (Not a Number) errors, effectively killing the training process.
To "police" these highways, DeepSeek implemented the Sinkhorn-Knopp algorithm. This 1967 procedure iteratively normalizes a matrix until it becomes doubly stochastic—meaning every row and every column sums exactly to 1.0.
By forcing the mixing behaviour of Hyper-Connections onto this mathematical manifold (known as the Birkhoff Polytope), DeepSeek achieved:
The mathematical core of this stability layer is derived from the following seminal work:
Sinkhorn, R., & Knopp, P. (1967). Concerning nonnegative matrices and doubly stochastic matrices. Pacific Journal of Mathematics, 21(2), 343-348.
In this paper, Sinkhorn and Knopp proved that any square matrix with strictly positive entries can be transformed into a doubly stochastic matrix by repeatedly scaling its rows and columns. While initially a problem of pure linear algebra, DeepSeek realized that this "Sinkhorn iteration" provides a perfect mechanism for Signal Normalization. By ensuring the mixing matrix $W$ satisfies $\sum_i W_{ij} = 1$ and $\sum_j W_{ij} = 1$, the network is prevented from adding artificial energy to the data stream, a requirement for training models with hundreds of layers.
The reason the Sinkhorn-Knopp iteration is so reliable for AI training is rooted in its mathematical proof of convergence. The proof essentially rests on the Total Support property.
This rigorous guarantee ensures that the "Manifold Constraint" in mHC isn't just a heuristic, but a mathematical certainty.
The Birkhoff Polytope is the set of all $n \times n$ doubly stochastic matrices. In the context of high-dimensional information, it functions as a geometric safe zone:
The stability provided by mHC enables the Internalized Chain of Thought (CoT). Traditionally, models perform reasoning by writing out steps in text. With mHC, researchers can stack hundreds of layers that act as internal reasoning modules. Because the signal remains stable, the model can perform multiple "logical passes" on information within its own internal layers before generating an answer.
Normalizing matrices thousands of times per second is typically too slow for industrial AI training. DeepSeek solved this through rigorous infrastructure optimization:
Industry analysts view the mHC paper as a technical preview for the rumoured DeepSeek-R2 flagship model, expected to launch around the Spring Festival in February 2026. DeepSeek-R2 was initially expected in 2025 but faced delays due to performance dissatisfaction and chip shortages. By implementing mHC, DeepSeek is expected to:
DeepSeek didn't just find a "patch"; they found a way to build a more complex "brain" that is mathematically guaranteed not to lose its mind during training. Looking back to 1967, they provided the structural integrity needed for the AI of 2026 to think more deeply, remain stable, and push the boundaries of machine reasoning.
This breakthrough provides a visual breakdown of how the Sinkhorn-Knopp algorithm acts as a safety rail, preventing signal explosion in the deep neural networks of the future. This DeepSeek mHC architecture explanation provides a high-level visual summary of how these mathematical manifolds facilitate smooth information flow across complex neural pathways.
The application of 1967 mathematics to the AI landscape of 2026 represents a profound turning point in the quest for Artificial General Intelligence (AGI). By reaching back to the Sinkhorn-Knopp algorithm, researchers have effectively solved the "structural fragility" that once capped the intellectual growth of neural networks.
This synthesis of mid-century linear algebra and modern GPU engineering has done more than stabilize training; it has granted models a "permanent internal logic". In 2026, the path to AGI is no longer just about adding more data or more power; it is about the mathematical elegance of equilibrium. The Sinkhorn-Knopp algorithm has become the stabilizer for a new era of "Internalized Reasoning," proving that the blueprints for our most advanced future minds were already written decades ago in the pages of pure mathematics.
Implementation Resources:
The complete Python implementation of the execution logic for both PyTorch and JAX, projecting matrices onto the Birkhoff Polytope manifold as detailed in this research, is available on GitHub.
This visual explanation of DeepSeek's mHC architecture summarizes how these mathematical manifolds facilitate deeper "thinking streams" in modern Transformers.
Tags: Agentic AI, AGI, Generative AI
 Glimpses of Agentic Intelligence: Gemini-3-Flash Navigating Mock ARC-AGI-3 Grid Worlds
Glimpses of Agentic Intelligence: Gemini-3-Flash Navigating Mock ARC-AGI-3 Grid Worlds
As of late 2025, the pursuit of artificial general intelligence (AGI) remains one of the most profound challenges in computer science. The ARC Prize Foundation, the steward of the Abstraction and Reasoning Corpus (ARC-AGI) benchmark, has steadily refined its evaluations to expose the limitations of current AI systems. While ARC-AGI-1 and ARC-AGI-2 focused on static visual puzzles that test core abstraction and reasoning—tasks humans solve near-perfectly but AI struggles with—the forthcoming ARC-AGI-3, slated for full release in early 2026, introduces a paradigm shift: interactive reasoning in dynamic, game-like environments. These environments demand exploration, planning, adaptation, and goal-directed behaviour over extended trajectories, qualities essential for human-like intelligence but elusive in today's models.
In anticipation of this benchmark, community-created demonstrations have emerged that simulate simplified ARC-AGI-3-style tasks. Two Jupyter notebooks—ARC_AGI_3_DEMO_case10.ipynb(10x10 grid) and ARC_AGI_3_DEMO_case64.ipynb (64x64 grid)—provide compelling offline proofs-of-concept. Both employ Google's newly released Gemini-3-Flash model (preview version, launched in December 2025) as an agent to solve a classic pathfinding problem: navigating a player (colour 1, blue) from a starting position to a goal (colour 2, red) while avoiding walls (colour 5, gray) on a grid. Actions are discrete (up, down, left, right), with collision detection and a win condition upon reaching the goal.
The smaller 10x10 demo features a compact maze: the player starts at [8,1] (near bottom-left), the goal at [1,8] (near top-right), and a horizontal wall barrier in row 4 (columns 2–7). The Manhattan distance—the theoretical minimum steps—is 14. Gemini-3-Flash solves it flawlessly in exactly 14 turns, achieving 100% action efficiency and zero collisions. This demonstrates optimal planning: the agent reasons about the obstacle, detours efficiently, and executes a shortest-path route without backtracking or errors.
Scaling up dramatically, the 64x64 demo places the player at [59,5] (near bottom-left) and goal at [5,59] (near top-right), with a near-complete horizontal wall at row 32 (midpoint) featuring a single gap at column 32. The optimal Manhattan distance balloons to 108 steps. Remarkably, Gemini-3-Flash again achieves perfection: completion in 108 turns, 100% efficiency, and zero collisions. The agent discovers the lone passage through exploration and reasoning, then navigates vast empty spaces with precision, showcasing robust spatial awareness over long horizons.
These results are striking for several reasons. First, they highlight Gemini-3-Flash's strengths in multimodal reasoning and agentic behaviour. The model receives the full grid as text (an extensive 2D list), recent action history, and a simple prompt: "Move 1 to 2. Avoid 5." It outputs structured JSON with a thought trace and action, leveraging high-level thinking modes to plan. In both cases, the agent avoids naive greedy moves (e.g., heading straight into walls) and exhibits foresight—essential for interactive benchmarks where trial-and-error alone would be inefficient.
Second, the flawless performance on optimal paths underscores emerging capabilities in spatial intelligence and obstacle avoidance, even in scaled environments. The 64x64 grid, with its sparse but critical obstacle, mimics the "novel unseen environments" ARC-AGI-3 aims to test: agents must generalize rules (movement, collisions) and adapt without prior training on identical layouts.
Yet, these demos also reveal the benchmarks' intent to probe deeper gaps. The tasks, while interactive, remain highly structured—deterministic physics, discrete actions, and clear goals—far simpler than the hundreds of diverse games planned for the full ARC-AGI-3, which will involve richer mechanics, longer horizons, and skill acquisition from scratch. Current frontier models excel in controlled simulations but often falter in true novelty, as evidenced by ongoing struggles on ARC-AGI-2 (top scores around 50-54% in late 2025). The perfect solves here suggest Gemini-3-Flash is a strong contender for early ARC-AGI-3 previews. Still, they also preview the humbling challenges ahead: humans would solve these intuitively and enjoyably, often faster or with creative shortcuts.
These notebooks, built on open repositories and leveraging accessible tools like Matplotlib for visualization, democratize experimentation with agentic AI. They offer a tantalizing preview of progress toward interactive reasoning—a cornerstone of AGI. As ARC-AGI-3 approaches, such demonstrations remind us that while models like Gemini-3-Flash are closing gaps in planning and navigation, the road to systems that learn and adapt as fluidly as humans remains long and exciting. They fuel optimism: with continued innovation, the agentic era may soon yield breakthroughs that redefine intelligence measurement itself.
Tags: Agentic AI, Generative AI, Predictive Analytics
 The Fusion of Perception and Reasoning: An AGI Approach to Aviation Safety via V-JEPA 2 with Gemini 3 Flash
The Fusion of Perception and Reasoning: An AGI Approach to Aviation Safety via V-JEPA 2 with Gemini 3 Flash
The history of aviation is defined by humanity's relentless pursuit of conquering the skies. This journey began with the daring ambition of the Wright brothers and the mythological warnings of Icarus. For over a century, safety in the air was bought with the hard-earned lessons of the past—often written in the aftermath of tragedy. However, we are entering a new epoch where we no longer need to wait for failure to learn. We are moving from a world of "reactive mechanics" to "proactive intelligence." This transition is fueled by the realization that proper safety lies not just in the strength of the steel but in the depth of the understanding. Today, we harness Artificial General Intelligence (AGI) to act as a digital sentinel, a vigilant mind that never tires and sees the very "DNA" of flight. By marrying the raw physics of motion with the high-level reasoning of human logic, we are fulfilling the ultimate promise of aviation: a sky that is not only accessible but inherently safe.
The foundation of this system is the Video Joint-Embedding Predictive Architecture (V-JEPA 2), which serves as the "sensory cortex" of the AGI. Unlike standard AI, which relies on static labels to identify objects, V-JEPA 2 is a predictive world model. It processes raw video of flight maneuvers—specifically landing sequences—and compresses them into a 1024-dimensional "Global Signature".
This signature represents the "physical DNA" of the flight, capturing the intricate relationship between mass, velocity, and gravity. Instead of looking for pixel patterns, the model understands the aircraft's motion in terms of Newtonian mechanics. The system calculates a Latent Prediction Error (LPE), a "surprisal" metric that quantifies how much the actual flight path deviates from a physically ideal landing. A high LPE score serves as an immediate red flag for potential safety violations.
While V-JEPA 2 provides the sensory data, the Gemini 3 model acts as the "prefrontal cortex," providing high-level reasoning. The integration of these two models allows the system to move beyond simple pattern matching into autonomous deliberation. Gemini receives the numerical "DNA" and LPE scores and interprets them using its vast internal knowledge base.
In a hard-landing scenario, Gemini does not just label the event; it reasons through the physics. It can distinguish between a "firm" but safe landing—where the airframe successfully transitions from aerodynamic lift to ground reaction mechanics—and a catastrophic failure where physical laws are violated. This capability allows the AGI to provide a transparent "verdict" rather than an opaque score.
Integrating Gemini 3 Flash with Meta's V-JEPA 2 creates a powerful "sensory-cognitive" loop, combining specialized physical world modelling with high-speed, frontier-level reasoning.
V-JEPA 2 (Video Joint Embedding Predictive Architecture) serves as the "eyes" of the system, trained on over a million hours of raw video to understand the laws of physics without human labelling.
Gemini 3 Flash serves as the decision-maker, processing abstract physical data from V-JEPA 2 to produce human-understandable logic and planning.
When these models are integrated, the resulting AGI (Artificial General Intelligence) pipeline can perceive, reason, and act within complex environments:
This video provides a deep dive into the original JEPA architecture and how V-JEPA uses latent representation prediction as its core objective to learn visual representations from video.
A critical new dimension of this AGI integration is its potential for Long-Term Structural Health Monitoring. Because the "Physical DNA" captures high-fidelity energy signatures of every landing, the agent can track the cumulative stress placed on an aircraft's airframe and landing gear.
By comparing the "Physical DNA" of multiple flights over time, Gemini can identify subtle shifts in an aircraft's response to impact—essentially detecting structural fatigue before it becomes visible to the naked eye. If the LPE during a landing is within nominal bounds but the "vibration signature" in the 1024-dimensional vector begins to shift from the baseline, the AGI can infer a loss of structural rigidity or dampening efficiency. This transforms the AGI from a real-time monitor into a predictive maintenance engine, ensuring safety is managed throughout the asset's lifecycle.
To understand where exactly a landing becomes "critical," the system generates a Surprise Score Profile. This graph plots the LPE over the duration of the landing sequence.
In a nominal landing, the surprise score remains low and stable as the plane descends, with only a predictable minor rise at touchdown. However, in a hard landing, the graph shows a sudden, sharp spike—like the 3.02 score observed in the demo—at the exact millisecond the landing gear strikes the runway. This visual "heartbeat" of the flight provides immediate, actionable evidence for safety investigators.
The model detects whether the airplane is landing and further categorizes the landing type. The system identifies the flight status through a multi-layered analysis:
The integration of V-JEPA 2 and Gemini 3 marks a paradigm shift in aviation safety, transitioning from reactive telemetry to proactive physical understanding. By moving beyond simple pixel recognition and instead capturing the "Physical DNA" of flight, this AGI framework enables a "digital twin" of Newtonian reality that can detect anomalies with unprecedented precision.
Key Technological Milestones
A New Era of Safety
The ultimate takeaway of this demo is that aviation safety no longer relies solely on human observation or binary sensor data. We are entering an era where Autonomous Safety Agents can "think" through the physics of a flight maneuver in real-time, providing a transparent, auditable, and physically grounded layer of protection for every asset in the sky. This convergence of computer vision and high-level reasoning doesn't just monitor flight—it understands it.
Tags: Agentic AI, AGI, Generative AI
 The Silicon Scientist: Gemini 3 Flash, High-Reasoning Agentic AI, and the Legacy of the Bose–Einstein Condensate
The Silicon Scientist: Gemini 3 Flash, High-Reasoning Agentic AI, and the Legacy of the Bose–Einstein Condensate
In 1924, Satyendra Nath Bose fundamentally altered the course of physics by describing a world where particles with integer spin—bosons—could overlap to form a single, coherent "super-atom." This state of matter, the Bose–Einstein Condensate (BEC), remained a theoretical prediction for 71 years until experimentalists finally achieved the required nanokelvin temperatures in 1995. Today, we are entering a third era of this legacy: one in which the observer is no longer just a human physicist but an Agentic AI capable of reasoning about the complex visual signatures of quantum matter.
The current implementation of a BEC simulation integrated with Gemini 3 Flash demonstrates a profound shift in scientific discovery. By combining a physics-based simulation with a "High Reasoning" AI agent, we create a closed-loop system where the machine generates data, visualizes it, and applies "Chain of Thought" reasoning to validate physical laws.
1. The Virtual Laboratory: Simulating the "Spike"
The simulation environment mimics the cooling of a boson gas. At high temperatures ($1.0\text{K}$), the system follows classical Maxwell–Boltzmann statistics, producing a broad, unimodal Gaussian distribution in its momentum space. As the simulation "cools" the system toward absolute zero ($0.01\text{K}$), it triggers the phase transition predicted by Bose: a macroscopic fraction of particles suddenly occupies the lowest-energy state. Visually, this is captured in a momentum histogram as a bimodal distribution—a sharp, high-density central spike sitting atop a broad thermal "pedestal."
2. The Architecture of Discovery: A Deep Dive into the Agentic BEC Simulation
The implementation of this demo is not merely a script but a closed-loop agentic ecosystem. It bridges the gap between classical numerical simulation and modern "High Reasoning" AI.
I. Physics Engine: The Stochastic Modelling of Bosons
The core of the simulation lies in the generate_bec_visual(temp) function, which uses the numpy library to model momentum distribution:
II. Multimodal Data Pipeline: In-Memory Visualization
To maintain a high-speed workflow, the system avoids the bottleneck of local file storage:
III. The Reasoning Agent: Gemini 3 Flash "High" Level
The most critical component is the call to the Gemini 3 Flash API using high-level reasoning configurations:
3. Results: Observed Simulation Phases
Based on the integrated simulation and analysis files, the following states were successfully identified:
Core Objective: The project demonstrates an agentic scientific workflow using Gemini 3 Flash to bridge the gap between numerical simulation and high-level physical reasoning
|
Phase |
Temperature |
Agent Observation |
Scientific Verdict |
|
Normal Gas |
1.0K |
Unimodal, broad Gaussian distribution (Maxwell-Boltzmann). |
No BEC formed. |
|
Critical Region |
0.1K |
Emergence of a bimodal distribution; onset of ground-state occupation. |
BEC formed. |
|
Condensate |
0.01K |
Distinct, sharp central spike sitting on a broad thermal "pedestal". |
BEC formation confirmed. |
Key Agentic Insights:
4. Conclusion: The Impact of Gemini 3 Flash on Scientific Discovery
The integration of Gemini 3 Flash into the analysis of Bose–Einstein condensates (BEC) represents a transformative leap in scientific communication and discovery. This agentic implementation proves that AI has evolved from a passive "helper" into an active "scientific supervisor," capable of bridging the gap between raw numerical data and theoretical grounding.
The project demonstrates that Gemini 3 Flash can deliver PhD-level reasoning while maintaining high-speed throughput. In the context of the BEC simulation, this enables real-time detection of complex quantum phase transitions—identifying the "bimodal signature" of a condensate within seconds—a task that historically required human experts to verify manually.
The true impact lies in the model’s native multimodality. By analyzing visual histograms directly from an in-memory buffer, the agentic AI bypasses the need for manual data stitching and visual artifact correction. It correctly identifies the macroscopic ground-state occupation predicted by Satyendra Nath Bose, not just through temperature readings, but through spatial pattern recognition of the "central spike" atop the thermal cloud.
As we approach the centenary of Bose's groundbreaking work, this demo serves as a modern tribute to his statistical genius. Bose reimagined the universe by discarding the distinct identities of microscopic particles, a philosophical leap that gave rise to quantum statistics. Today, agentic AI like Gemini 3 Flash honours this legacy by automating the verification of his theories, grounding its "Scientific Verdicts" in the very indistinguishability and wave-overlap principles Bose first described.
In the legacy of Satyendra Nath Bose, we are no longer just looking at the universe; we are teaching our machines to understand and explain the deep, underlying beauty of its quantum order.
Satyendra Nath Bose: The Collaborator Who Gave Birth to Bose-Einstein Statistics!
Tags: Generative AI, Agentic AI, AGI
 World Models: The Foundational Architecture for Artificial General Intelligence
World Models: The Foundational Architecture for Artificial General Intelligence
The pursuit of Artificial General Intelligence (AGI)—systems capable of learning, understanding, and applying intelligence across diverse tasks like a human—is hampered by a fundamental flaw in current AI architectures. Contemporary deep learning models, while exhibiting spectacular performance in narrow domains, are overwhelmingly data inefficient, often requiring millions of examples to learn what a child grasps in one or two. Furthermore, they struggle with causality and long-horizon planning, operating primarily as powerful, yet reactive, pattern matchers. The solution lies in a cognitive architecture that mirrors the human brain's most powerful feature: the ability to imagine. This architecture is the World Model. Far from being merely a robotics tool, World Models represent the most promising paradigm shift toward AGI, fundamentally by teaching AI systems the basic, causal, and common-sense principles of the world, whether physical, biological, or digital.
The concept of an internal model for predicting the future is not a new invention but an evolutionary convergence of ideas from psychology, control theory, and machine learning.
The philosophical foundation of World Models lies in theories of human cognition. As early as the 1940s, a prominent psychologist proposed that the human mind builds "small-scale models" of external reality to anticipate events and "try out various alternatives" before taking action. This concept—that the brain acts as an internal simulator—is the psychological ancestor of the computational World Model. Concurrently, in engineering, model-based control became standard. This approach, encapsulated in the Good Regulator Theorem (which states that every good regulator of a system must be a model of that system), relied on explicit mathematical models of a plant's dynamics to calculate control signals, establishing the mathematical necessity of an internal system model for adequate control.
The transition to modern AI began when researchers sought to merge the explicit models of control theory with the learning capabilities of early machine learning. In 1990, the Dyna architecture was proposed, representing one of the earliest explicit integrations of planning and Reinforcement Learning (RL). Dyna agents used real-world experience to train a simple transition model, which was then used to generate simulated experience (planning in imagination) to train the agent's policy further. This was a crucial shift, demonstrating that simulated experience could accelerate real-world learning and directly prefiguring the sample-efficiency argument. Pre-deep learning approaches, however, were limited because their models relied on hand-crafted state features, making them too brittle to handle the complexity of raw sensory data, such as pixels.
The breakthrough arrived when deep neural networks provided the tools to manage high-dimensional inputs. The seminal "World Models" paper formalized the modern concept. The key innovation was using deep learning architectures (such as Variational Autoencoders) to address the perception problem: the Encoder Model compressed raw pixels into a low-dimensional latent space. This allowed the Dynamics Model to efficiently predict the future in this abstract, computationally efficient space. Subsequent advancements in algorithms established latent imagination as the state of the art for continuous control. Today, this concept is scaling to foundation models (such as those used for text-to-video generation), which are widely viewed as powerful, generative World Models that learn physics from video data, cementing the architecture as the core cognitive piece required for general intelligence.
Having established its historical roots, the World Model's first modern contribution is to address the sample-efficiency crisis plaguing Model-Free Reinforcement Learning (RL). Traditional RL agents learn through massive trial-and-error, directly mapping sensory inputs to actions based on accumulated reward. This methodology is impossibly slow and resource-intensive for real-world applications, proving infeasible for tasks that require physical interaction or long training cycles. World Models resolve this by functioning as a generative internal simulator. The system first learns an Encoder Model to compress high-dimensional raw inputs (like video frames) into a concise, low-dimensional latent space. Crucially, the Dynamics Model is then trained to predict the next latent state from the current state and the chosen action. This enables the agent to perform latent-space planning—or "dreaming"—by running forward simulations entirely within the model, generating synthetic experience data at extremely high speed. In applications like Game AI, agents can accrue millions of virtual interactions, accelerating learning and achieving far greater sample efficiency than their real-world counterparts. This ability to learn from imagination rather than constant real-world interaction is a non-negotiable step toward AGI.
A second failure of reactive AI is its inability to perform long-horizon planning—the capacity to sequence dozens of steps to achieve a distant goal—and to ensure safety through foresight. Reactive systems select the best immediate action based on the current state. World Models imbue the agent with accurate temporal foresight and causal understanding. By using its internal Dynamics Model, the agent can perform counterfactual reasoning: it can simulate multiple possible futures resulting from different initial actions and evaluate which sequence maximizes the long-term expected reward. This is essential for safety-critical non-robotics applications. For instance, in Autonomous Vehicles (AVs), the World Model is used not just to classify objects, but to predict the trajectories of all surrounding vehicles and pedestrians over the next five seconds. This allows the system to test a potentially risky maneuver (e.g., a lane change) in simulation and predict a catastrophic outcome (a crash) before executing it in reality, making the system safer and more deliberative.
Modelling Complex, Generalized Digital Dynamics
The significance of World Models extends beyond the domain of physical reality to any system governed by complex, high-dimensional dynamic principles. The goal of AGI is to generalize, and World Models are the architecture for learning generalized dynamics—traditional, equation-based modelling struggles with the non-linear, chaotic nature of systems like climate or financial markets. World Models, however, are trained to find the underlying dynamical principles of any system, regardless of its domain. They are purely statistical models that learn the flow of complex data. This has far-reaching applications in Climate modelling and Forecasting. By training World Models on massive datasets of satellite imagery and atmospheric sensor readings, systems learn the physics of the atmosphere and oceans, providing more accurate, physics-consistent, and high-resolution forecasts than older methods. Similarly, dynamic network systems (traffic, supply chains, economics) can be modelled. By succeeding in these diverse, non-physical domains, World Models demonstrate their fundamental nature as a general-purpose cognitive tool, capable of abstracting and predicting the rules of any complex system.
Finally, World Models provide the crucial link that currently separates powerful Large Language Models (LLMs) from achieving AGI: grounding and common sense. While LLMs are masters of linguistic reasoning, they are essentially "brains floating in linguistic space," lacking an understanding of the physical consequences of the words they use (e.g., gravity, friction, object permanence). A World Model, particularly one trained on massive amounts of video and sensorimotor data (a Vision-Language-Action, or VLA, foundation), learns the intuitive physics of the world purely through observation. This provides the causal framework—the "rules of reality"—that an LLM can reference. A complete AGI will likely use the LLM for high-level, symbolic reasoning and planning, while delegating the physical plausibility checks to the World Model. This integration solves the Reality Gap and transforms symbolic reasoning into physically grounded action, ensuring that abstract plans are causally coherent and robust against unexpected real-world events.
The advancement of AI towards AGI necessitates a cognitive architecture that transcends simple pattern matching. World Models deliver on this necessity by implementing an internal simulator capable of imagination and foresight. They are the mechanism that provides four essential capabilities for general intelligence: radical sample efficiency through dreaming, robust long-horizon planning via counterfactual reasoning, generalized modelling across diverse dynamic systems, and the grounding of language in physical reality. By moving AI from reactive systems to predictive, deliberative agents, World Models are not just improving existing technology—they are realizing the historical convergence of cognitive theory and engineering by constructing the necessary cognitive backbone that will define the next generation of generally intelligent machines.
Tags: Agentic AI, AGI, Generative AI
 The Agentic Superiority of Gemini 3 Pro: Scale, Multimodality, and Ecosystem Integration
The Agentic Superiority of Gemini 3 Pro: Scale, Multimodality, and Ecosystem Integration
The contest between Google's Gemini 3 Pro and OpenAI's GPT-5.2 marks the pinnacle of modern AI capability. Still, in the specific domain of agentic workflows—the ability to reliably perform multi-step, tool-using, and state-retaining tasks—Gemini 3 Pro demonstrates a distinct and strategically valuable advantage. While GPT-5.2 excels in raw abstract reasoning and structured coding benchmarks, Gemini 3 Pro is architected for the sheer scale, multimodal complexity, and seamless integration required by true autonomous agents operating in the enterprise environment.
The foundational strength of Gemini 3 Pro for agentic tasks is its unprecedented context window of up to one million tokens. An AI agent, by definition, must maintain a memory of its instructions, a log of its past actions, the output of external tools, and the data it is currently analyzing. GPT-5.2's significant 400k-token capacity is formidable, but Gemini 3 Pro's 1M-token window translates directly into superior state retention and long-horizon planning stability. An agent tasked with analyzing a complete software repository, a year's worth of financial reports, or a lengthy legal contract can ingest the entire corpus in a single call. This eliminates the need for complex, error-prone Retrieval-Augmented Generation (RAG) chunking or arbitrary truncation, reducing "reasoning drift" and ensuring the agent's decisions are based on a holistic, fully-aware view of the entire operational context.
Furthermore, agentic work in the real world is inherently multimodal. A business agent may be asked to "analyze the Q3 sales video transcript, compare the figures against the attached spreadsheet image, and update the quarterly report." Gemini 3 Pro's state-of-the-art native multimodality gives it a potent edge here. It is built to process and reason across text, images, video, and audio simultaneously. While GPT-5.2 has made significant advances in vision, Gemini 3 Pro's strength in complex visual and spatial reasoning, particularly in interpreting dense charts, graphs, and unstructured documents, provides a richer, more accurate input foundation for agent decision-making.
Finally, the agentic advantage of Gemini 3 Pro is secured by its deep integration within the Google ecosystem. An agent is only as good as the tools it can reliably wield. Gemini 3 Pro is designed to function as the core orchestrator within Google Workspace, enabling direct, high-fidelity interaction with Google Docs, Sheets, and Calendar. For the vast number of businesses and developers operating within this ecosystem, Gemini 3 Pro offers ready-made, production-grade workflows for tasks such as automating report generation, financial modelling, and supply chain adjustments. Google's development of agentic platforms and tools further accelerates this advantage, positioning Gemini 3 Pro as the preferred brain for autonomous enterprise automation.
The assumption that one model is inherently "smarter" is often misleading; models excel at different types of reasoning that require distinct computational approaches. Gemini 3 Pro's Deep Think is an enhanced mode that instructs the model to explore a broader range of possibilities, while GPT-5.2's top tiers are tuned for predictable, structured execution.
| Reasoning Metric | GPT-5.2 (Pro/Thinking) | Gemini 3 Deep Think | Winner / Characteristic |
| Abstract Visual Reasoning (ARC-AGI-2) | ~54.2% | ~45.1% | GPT-5.2 (Stronger in non-verbal, fluid intelligence puzzles.) |
| Graduate-Level Science (GPQA Diamond) | ~93.2% | ~93.8% | Gemini 3 Deep Think (Slightly better on complex scientific knowledge/theory.) |
| High School Math (AIME 2025) | 100% (No tools) | 95.0% (No tools) / 100% (With tools) | GPT-5.2 (Better raw mathematical logic without external tools.) |
| Theoretical Reasoning (Humanity's Last Exam) | ~34.5% | ~41.0% | Gemini 3 Deep Think (Excels in open-ended, theoretical physics/philosophy.) |
| Execution Reliability | Stronger | Highly capable, but higher latency. | GPT-5.2 (Optimized for predictable, consistent automation/tool use.) |
Gemini 3 Deep Think focuses on theoretical depth and scientific understanding. It builds a broader array of internal reasoning paths, exploring multiple hypotheses before settling on a solution. This makes it highly effective in abstract and scientific research environments, scoring marginally higher on tests like GPQA Diamond and significantly higher on Humanity's Last Exam.
GPT-5.2's core is tuned for structured reasoning and reliable execution in professional workflows. It shows a clear advantage on benchmarks like ARC-AGI-2, which measures fluid intelligence and the ability to solve abstract, novel, non-verbal problems. This translates into superior general-purpose problem decomposition and a more predictable, reliable agent for deployment where execution errors are costly.
In conclusion, while GPT-5.2's remarkable abstract reasoning and high scores on specific coding benchmarks provide a crucial intellectual core, the practical demands of autonomy—massive context memory, complex multimodal input, and seamless tool execution—tip the scales toward Gemini 3 Pro. Its architecture is explicitly designed to move beyond singular brilliance to achieve reliable, persistent, multi-step action at a scale unmatched by its contemporary, solidifying its position as the stronger foundational model for the next generation of AI agents.
The choice between these two powerful models for agentic deployment often comes down to the specific environment and the nature of the task. Gemini 3 Pro offers advantages for scale and integration, while GPT-5.2 leads in pure reasoning complexity
| If your agentic workflow is... | Choose Gemini 3 Pro | Choose GPT-5.2 |
| Focused on Data/Documents/Visuals | YES. Analyzing a 500-page PDF with charts or managing a multi-tab Google Sheet. | Maybe. Good for analyzing text, but Gemini is richer for visual/spatial data. |
| Heavily Integrated with Google | YES. Automating tasks across Gmail, Docs, or Calendar. | No. Requires external connectors (e.g., Zapier), which adds complexity. |
| Complex Reasoning/Coding | Maybe. Excellent memory for codebases, but GPT-5.2 leads on hard-coding benchmarks (SWE-Bench Pro). | YES. For self-debugging, large-scale refactoring, or breakthrough problem-solving. |
| Needs Maximum State Memory | YES. Its 1M-token context gives it the most reliable long-term memory for an ongoing task. | No. Max 400k tokens. |
Tags: Generative AI, Agentic AI, AGI
 The New Silicon Frontier: Specialization and the Diverse Landscape of AI Chips
The New Silicon Frontier: Specialization and the Diverse Landscape of AI Chips
The rapid ascension of Artificial Intelligence, from nascent deep learning models to today's gargantuan generative AI systems, has been wholly dependent on a parallel revolution in hardware. General-purpose Central Processing Units (CPUs), designed for sequential tasks, quickly became bottlenecks for the massive, highly parallel computations inherent in neural networks. This necessity has forged a new silicon frontier, resulting in a diverse and highly specialized landscape of AI accelerators—chips purpose-built to execute AI workloads with unprecedented speed, efficiency, and scale.
The competitive landscape is best understood through the architectural core and primary role of each chip type:
|
Chip Category |
Specific Chip Example |
Primary AI Role(s) |
Architectural Core |
Key Optimization/Feature |
| GPU |
NVIDIA H100, AMD Instinct |
Model Training & High-Performance Inference |
Thousands of Parallel Streaming Multiprocessors (SMs) / Compute Units |
High Memory Bandwidth (HBM), General Purpose Parallelism (CUDA/ROCm) |
|
ASIC (Cloud - Training) |
AWS Trainium |
Model Training |
Proprietary NeuronCores with massive on-chip SRAM |
Cost-effective Training at Scale, Distributed Architecture (NeuronLink) |
|
ASIC (Cloud - General) |
Google TPU |
Model Training & Inference |
Systolic Array of Matrix Multipliers (MAC units) |
Unmatched Performance-per-Watt for Tensor-based operations (TensorFlow/JAX) |
|
ASIC (Cloud - Inference) |
AWS Inferentia |
Model Inference |
Proprietary NeuronCores optimized for low latency |
Lowest cost per inference, high throughput, minimized data movement. |
|
ASIC (Edge/Mobile NPU) |
Apple Neural Engine |
Model Inference |
Specialized Inference Accelerators (Varies by generation) |
Extreme Power Efficiency, On-device processing for privacy and low latency. |
|
FPGA |
Intel Stratix, AMD Versal |
Real-time Inference & Signal Processing |
Reconfigurable Logic Blocks (LUTs) and Dedicated Multipliers |
Hardware Reconfigurability, Deterministic Latency, Customizable Data Paths. |
The fundamental differences in AI hardware stem from their core architectural designs, which determine their suitability for either the energy-intensive training phase or the low-latency inference phase.
GPUs, exemplified by the NVIDIA H100, dominate large-scale AI training due to their fundamental design philosophy: massive parallelism. Unlike CPUs, which have a few powerful cores optimized for sequential instruction processing, GPUs have thousands of smaller, more efficient Streaming Multiprocessors (SMs).
ASICs represent the ultimate commitment to performance and efficiency for a fixed task, often achieving better performance per watt than GPUs.
The Systolic Array architecturally defines the TPU. This is a grid of interconnected Multiply-Accumulate (MAC) units where data (tensors) flows rhythmically, allowing hundreds of thousands of operations to co-occur while minimizing data movement and power consumption.
The ANE is a prime example of an NPU (Neural Processing Unit) for the edge. It is highly optimized for executing inference with minimal power draw, keeping AI processing on-device to enhance privacy and provide ultra-low latency.
FPGAs offer the unique ability to reconfigure their hardware logic after manufacturing via an array of Configurable Logic Blocks (CLBs). This allows FPGAs to achieve deterministic, ultra-low latency for real-time applications and provides a balance between the efficiency of an ASIC and the flexibility of a GPU.
While specialized chips drive efficiency gains per calculation, the overall environmental footprint of the hardware ecosystem is rapidly expanding. This ecological cost spans the entire lifecycle of the chip.
The most significant impact is the embodied carbon and pollution generated before use. Fabrication is extremely resource-intensive, requiring massive amounts of rare earth elements and water, and is energy-intensive, releasing highly potent greenhouse gases.
The immense performance of AI accelerators places massive operational demands on data centers.
The speed of the AI hardware arms race creates a severe e-waste problem. The competitive landscape pushes companies to replace high-performance components every few years, generating enormous volumes of electronic waste containing toxic substances like lead and mercury.
The key negative feedback loop in the AI industry: the relentless pursuit of performance directly drives a massive environmental problem.
To mitigate this environmental crisis, the industry is actively investing in next-generation thermal management and circularity models.
The adoption of sustainable solutions offers significant financial advantages, making green initiatives a strategic business imperative.
The circular economy model provides financial security:
The evolution of the AI chip is more than a story of technical progress; it is a critical narrative of specialization driven by immense computational demand. The future of intelligence is being sculpted in silicon, dictated by the efficiency of the Systolic Array, the throughput of the NeuronCore, and the high bandwidth of HBM memory.
Yet, this power comes with a profound price: the exponential ecological impact of embodied carbon, water consumption, and the rising tide of e-waste. This realization has forced the industry into a necessary, rapid convergence in which peak performance and sustainability are no longer mutually exclusive but mutually dependent.
The transition to efficient Direct-to-Chip and Immersion Cooling systems, coupled with ambitious Circular Economy programs for component reuse, is not merely an act of environmental stewardship. It is a strategic economic imperative. These initiatives yield direct financial benefits, secure supply chains, reduce operational costs, and meet the mandatory ESG requirements of global investors.
Ultimately, the choice of AI hardware has transcended engineering specifications. It is now a defining ethical and economic decision that determines not only the speed of the next generative model but the resilience of the planet's resources. The final frontier of AI is not conquering complexity, but mastering sustainability, ensuring that the relentless pursuit of intelligent machines does not come at the expense of a viable future.
Tags: Generative AI, Agentic AI, AGI
 The Hardware Foundation of Future AI: Tensor Processing Units, Agentic AI, and the Road to AGI
The Hardware Foundation of Future AI: Tensor Processing Units, Agentic AI, and the Road to AGI
The choice between a TPU and a Graphics Processing Unit (GPU) for AI workloads often comes down to a trade-off between specialization (TPU) and versatility (GPU). The TPU's role in future AI is best understood in comparison to its dominant competitor:
The pursuit of more sophisticated Artificial Intelligence, from multi-step Agentic AI to the eventual realization of Artificial General Intelligence (AGI), is fundamentally a pursuit of compute. At the heart of this drive is the Tensor Processing Unit (TPU), Google's custom-designed Application-Specific Integrated Circuit (ASIC). By trading the general-purpose flexibility of traditional CPUs and GPUs for extreme specialization in deep learning's linear algebra, TPUs have created the necessary infrastructure for training and deploying the massive models that underpin today's and tomorrow's most ambitious AI systems.
The core innovation of the TPU lies in its architecture, which is built around the systolic array. This design allows data, in the form of tensors (multidimensional arrays), to flow rhythmically through a grid of thousands of multiply-accumulate units. This highly optimized, assembly-line approach drastically reduces the need for constant, slow memory access, bypassing the classic von Neumann bottleneck that constrains general-purpose processors.
This architectural choice yields three critical benefits:
The choice between a TPU and a Graphics Processing Unit (GPU) for AI workloads often comes down to a trade-off between specialization (TPU) and versatility (GPU). The TPU's role in future AI is best understood in comparison to its dominant competitor:
| Feature | Tensor Processing Unit (TPU) | Graphics Processing Unit (GPU) |
| Design/Architecture | ASIC (Application-Specific Integrated Circuit). Uses a Systolic Array designed exclusively for dense matrix multiplication. | General-Purpose Processor. Uses thousands of programmable cores. |
| Primary Focus | Specialized for AI/ML. Optimized for tensor algebra, particularly for training and inference of large neural networks. | Versatile. Used for graphics rendering, scientific computing, and general AI/ML. |
| Energy Efficiency | Higher Performance per Watt for AI workloads. | Less efficient for dense matrix math, with higher overall power consumption per chip. |
| Flexibility | Limited. Optimized for specific frameworks (like TensorFlow and JAX). | High. Broad support for all major frameworks (PyTorch, TensorFlow, etc.) and custom operations. |
| Scalability | Designed for massive scale via TPU Pods (thousands of interconnected chips). | Scales well with interconnects but is generally limited to smaller clusters. |
For workloads that perfectly fit the deep learning model and use the optimized software stack, TPUs often offer significantly better performance per dollar and energy efficiency than contemporary GPUs. For specific workloads, such as large language model training, recent TPU generations have been shown to offer superior value. However, GPUs remain the industry standard for their unmatched flexibility and broad ecosystem, making them the preferred choice for researchers and tasks requiring custom operations or diverse computational needs. The ultimate trend is that TPUs are the powerhouses for achieving extreme scale in training frontier models, while GPUs maintain dominance through versatility and accessibility.
The specialized capabilities of TPU are crucial for advancing AI beyond its current state.
Agentic AI systems, which rely on AI agents to plan, execute multi-step workflows, and coordinate with tools, are directly enabled by TPU efficiency. TPUs accelerate the training and continuous fine-tuning of the competent foundation models that serve as the agents' cognitive core. Furthermore, for agentic workflows involving dozens or hundreds of sequential model calls, TPUs provide the high throughput and low latency necessary for cost-efficient inference at scale, making large fleets of active agents economically viable.
The realization of Artificial General Intelligence (AGI) is often framed as a problem of scale, requiring models exponentially larger than those available today. TPUs provide the maximum available computational fabric today through the TPU Pod architecture, enabling unprecedented numbers of parameters to capture the vast, interconnected knowledge and emergent reasoning abilities required for AGI. By drastically reducing the time needed to train a massive experimental model, TPUs accelerate the entire research pipeline—a vital process for exploring novel architectures and training techniques that may lead to an AGI breakthrough.
In conclusion, the TPU is more than just a fast chip; it is an economic and architectural blueprint for massive-scale, energy-efficient AI. It is the powerhouse that trains the large language models, enabling today's Agentic AI workflows and providing the essential compute density required to move closer to the era of AGI. Without this specialized hardware foundation, the current trajectory of rapid AI advancement would be severely constrained by the limitations of general-purpose computing.
Tags: Generative AI, Open Source, Agentic AI
 The AI Trilemma: Competition, Infrastructure, and the Acceleration of Agentic AI
The AI Trilemma: Competition, Infrastructure, and the Acceleration of Agentic AI
The artificial intelligence industry is currently defined by a hyper-competitive trilemma, where advances in model capability, infrastructure efficiency, and commercial viability interact to accelerate the path toward Artificial General Intelligence (AGI). The recent confluence of OpenAI’s internal “code red,” Amazon’s launch of the cost-disruptive Trainium3 chip, and Mistral AI’s release of the open-source, multimodal Mistral Large 3 model reveals that the race is no longer simply about building the biggest model, but about forging the modular, efficient, and reliable ecosystem required to deploy truly autonomous, agentic systems. This intense competitive pressure is forcing the industry to focus on the essential building blocks—efficiency and modularity—that must be solved before AGI can be realized.
The development of sophisticated Agentic AI—systems capable of autonomous planning, tool use, and long-term goal execution—is fundamentally dependent on model capability, a domain that Mistral AI’s latest release has significantly advanced. The Mistral Large 3 model, with its Sparse Mixture-of-Experts (MoE) architecture (featuring 41 billion active parameters in a forward pass from a 675 billion total pool), large 256K context window, and native multimodal (vision) and multilingual support across 40+ languages, provides the foundational intelligence required for multi-step tasks. Its instruction-tuned version has achieved parity with the strongest closed models and ranked #2 among open-source non-reasoning models, signalling world-class performance. Crucially, its Apache 2.0 open-source, permissive license democratizes access to this frontier capability, moving the development of advanced agents out of a few proprietary labs and into the broader developer community. Agentic systems thrive on tool use and structured outputs (like JSON); by baking superior function-calling capabilities into an efficient MoE model, Mistral delivers the intelligence at scale needed for complex decision-making while maintaining high operational efficiency. This innovation, complemented by the compact Ministral 3 family for edge deployment, is critical to AGI, as it is widely predicted to manifest not as a single monolithic model but as a network of highly specialized, interacting agents.
However, complex agent networks require massive, continuous computational power, making the economics of AI infrastructure the second, indispensable driver. Amazon’s announcement of the Trainium3 chip, promising up to 50% lower training and operating costs compared to existing GPUs, addresses the core financial obstacle to large-scale AI deployment. Built on 3-nanometer technology, the Trn3 UltraServers deliver over 4 times (4.4x) the compute performance and 40% greater energy efficiency than their predecessor, scaling up to a massive 144 chips per system. This performance, already being leveraged by key rivals like Anthropic for production workloads, makes the cost of AI development and inference radically cheaper. A single complex agent executing hundreds of intermediate thoughts, API calls, and long-range planning steps generates dramatically more inference usage than a simple, single-query chatbot. If AGI is to be built from thousands of simultaneously running agents, the cost of running those agents must approach zero. Trainium3, alongside Google's TPU efforts, challenges Nvidia's market dominance by creating a much-needed environment of cost competition. Most significantly, Amazon's strategic decision to have Trainium4 support Nvidia's NVLink Fusion interconnect technology is a pragmatic hedge, offering enterprises a path to diversify their hardware reliance without abandoning the dominant CUDA ecosystem entirely. The infrastructure war is, therefore, a quiet but profound accelerator of AGI’s deployment potential.
Finally, the competitive crisis at OpenAI highlights the essential need for core product reliability and usability—qualities that must precede any attempt to deploy AGI. Sam Altman's "code red" directive, redirecting resources away from new revenue initiatives (like shopping agents and ads) to focus entirely on improving ChatGPT's speed, reliability, and personalization, signals a crucial maturation in the industry. For Agentic AI to function in the real world (e.g., managing a budget or scheduling complex events), they cannot be slow, unreliable, or prone to catastrophic failure. An autonomous agent must be fundamentally trustworthy. The focus on improved personalization is also key, as AGI systems must be capable of maintaining long-term state, learning from cumulative interactions, and adapting their persona and output to individual users—a core requirement for any truly general intelligence. This "code red" is thus less a retreat and more a tactical prioritization of the stability and trust layers upon which any ambitious AGI project must be built.
In conclusion, the current landscape—marked by competitive urgency (OpenAI), infrastructure efficiency (Trainium3), and open innovation (Mistral Large 3)—is rapidly establishing the prerequisites for AGI. The fight for market share is driving down computational costs and forcing the development of specialized, efficient, and reliable models well-suited for agentic deployment. A single, sudden breakthrough in research may not define the path to AGI. Still, the gradual, competitive convergence of cost-effective, modular, and multimodal agentic building blocks is now being created and scaled at an unprecedented rate across the entire technology stack.
Tags: Agentic AI, Generative AI, Open Source
 The AI Curriculum: A Library's Deep Dive into Artificial Intelligence
The AI Curriculum: A Library's Deep Dive into Artificial Intelligence
The collection of 29 distinct audiobook titles focused on Artificial Intelligence, Machine Learning, and Deep Learning is not merely a library; it represents a comprehensive, multi-faceted curriculum covering the technical foundations, real-world applications, strategic business implications, and profound existential questions posed by modern AI. This concentration of titles demonstrates a dedicated pursuit of knowledge across the entire AI landscape, from foundational code to global, socio-political forecasting.
The most fundamental layer of this curriculum addresses the practical engineering and scientific principles required to build and understand AI systems. Titles like Build a Large Language Model (From Scratch), Deep Learning with Python (Second Edition), and Deep Learning with Python provide the hands-on, code-level knowledge necessary for model creation and implementation. These texts sit alongside broader architectural guides, such as Designing Machine Learning Systems: An Iterative Process for Production-Ready Applications and Grokking Artificial Intelligence Algorithms: Understand and Apply the Core Algorithms of Deep Learning and Artificial Intelligence in This Friendly Illustrated Guide, Including Exercises and Examples. These books collectively explore the internal mechanisms of intelligence, from the neural network structures to the principles of scalable deployment.
The technical focus extends to the practicalities of professional development, with titles such as The AI Engineering Bible: Guide to Build, Develop, and Scale Production-Ready AI Systems, Software Engineering at Google: Lessons Learned from Programming Over Time, and the specialized Clean Code: A Handbook of Agile Software Craftsmanship. These selections emphasize that successful AI development is inseparable from sound software engineering practices and architectural design, as detailed in Fundamentals of Software Architecture: An Engineering Approach.
Moving beyond the technical core, a significant portion of the collection explores how AI systems are deployed, managed, and monetized in the real world. This section is highly focused on business and strategic implementation. LLMs in Production: Engineering AI Applications and Generative AI in Practice: 100+ Amazing Ways Generative Artificial Intelligence Is Changing Business And Society directly address the immediate commercial impact of large language models and other generative techniques.
Agentic Artificial Intelligence highlights the shift towards autonomous AI behaviour: Harnessing AI Agents to Reinvent Business, Work and Life, suggesting a focus on the next evolution of automated systems. The theme of professional application is also evident in niche areas, such as the sector-specific book Artificial Intelligence in Healthcare: AI, Machine Learning, and Deep and Intelligent Medicine Simplified for Everyone, which demonstrates an interest in how AI is transforming traditional industries. Finally, for those looking to capitalize on this wave, the provocative title ChatGPT Become a Millionaire: Capture the AI ChatGPT Market and Become a Millionaire suggests an interest in the entrepreneurial potential of new AI tools.
The final, most compelling layer of this library is dedicated to the philosophical, political, and existential impact of advanced AI. This is where the curriculum expands into human-machine futures. Questions of global power are central to the geopolitical analyses in AI Superpowers: China, Silicon Valley, and the New World Order and The Coming Wave: AI, Power, and Our Future. These titles examine the race for technological dominance and the risks it poses to international stability.
Closer to the philosophical core of AI, titles like Life 3.0: Being Human in the Age of Artificial Intelligence, Superintelligence: Paths, Dangers, Strategies, and Human Compatible: Artificial Intelligence and the Problem of Control directly tackle the "control problem"—the risks associated with creating intelligence greater than our own. These are balanced by works that frame intelligence and consciousness, such as A Thousand Brains: A New Theory of Intelligence and Being You: A New Science of Consciousness.
The chronological and societal impact is framed by AI 2041: Ten Visions for Our Future, offering a view of the near-term future, while Genesis: Artificial Intelligence, Hope, and the Human Spirit provides a deeper reflection on the spiritual and humanitarian implications. Even historical context is provided by titles like Nexus: A Brief History of Information Networks from the Stone Age to AI and The Deep Learning Revolution, showing the continuity of information processing from the ancient world to the present.
This curated selection of 29 audiobooks forms an unparalleled personal curriculum, mapping the intellectual landscape of Artificial Intelligence from machine code to human consciousness. By engaging with works on deep learning, architectural design, business strategy, global politics, and existential philosophy, this library reflects a profound commitment not just to understanding AI as a technology but as a defining force shaping the future of humanity.
Tags: Agentic AI, Generative AI, Open Source
 The Modular Ascent: Integrating Gemini 3, V-JEPA, and World Models for Aviation AGI
The Modular Ascent: Integrating Gemini 3, V-JEPA, and World Models for Aviation AGI
The dream of Artificial General Intelligence (AGI)—a machine capable of matching human cognitive flexibility—has driven computer science since the Dartmouth Workshop in 1956. For decades, this pursuit was divided: the Symbolic AI tradition focused on formal rules and logic, often failing to interface with the messy, continuous real world; simultaneously, the Connectionist (Deep Learning) tradition excelled at perception and pattern recognition but lacked intrinsic causality and high-level reasoning. The advent of powerful Large Language Models (LLMs) like Gemini, with their vast store of codified human knowledge, reignited the AGI debate but highlighted a persistent gap: how does a text-based brain effectively govern a body in the physical world?
This work directly tackles that gap. Inspired by the architectural pillars proposed by influential thinkers such as Yann LeCun, the system presented here demonstrates true modularity. It transcends the limitations of monolithic LLMs by integrating Vision Joint-Embedding Predictive Architecture (V-JEPA) for real-world sensing, a Predictive Latent Dynamics Model (PLDM) for internal causal simulation, and the advanced reasoning of Gemini 3 Pro for operational oversight. By combining these specialized modules, the architecture aligns with the five core AGI pillars, resulting in a unified, agentic system capable of coherent action in a complex environment such as autonomous flight operations. This integration represents a critical evolutionary leap from abstract knowledge processing toward embodied, causal, and safe decision-making.
The successful refactoring of the code showcases the integration of an LLM (Gemini 3) with a perception system (V-JEPA) and a dynamics model (PLDM) to conceptually demonstrate the Five AGI Pillars for an autonomous flight agent. The entire notebook structure—from data ingestion and model training to the final Gemini assessment—is designed to address these fundamental requirements of next-generation AI.
Pillar Alignment: The system explicitly uses a Latent Dynamics Predictor (the "World Model") to learn the causal relationships of aircraft states in a hidden, compact space. Code Implementation:
Pillar Alignment: The system moves beyond memorizing patterns by building a predictive model that understands cause-and-effect ($\text{Action} \to \text{Next State}$) in the latent space. Code Implementation:
Pillar Alignment: The system is inherently modular, separating Perception (V-JEPA for feature extraction), High-Level Reasoning (Gemini LLM for operational assessment), and Causal Planning (Latent Dynamics Predictor). Code Implementation:
Pillar Alignment: The agent is embodied through its visual input (V-JEPA processing a video from an assumed aircraft perspective) and its reliance on physical state data (ADS-B telemetry). It focuses on what matters—the operational context. Code Implementation:
Pillar Alignment: This describes a hybrid system, demonstrated here by combining the mathematically rigorous Cognitive World Model (the latent state predictor) with a Symbolic Reasoning system (the Gemini LLM). Code Implementation:
The architecture demonstrated by integrating V-JEPA, the Predictive Latent Dynamics Model, and Gemini 3 Pro's advanced reasoning represents a pivotal shift from narrow AI utility to the design of truly agentic AGI systems. The success of this modular approach validates the need to combine specialized components: V-JEPA for what is seen, PLDM for what will happen, and Gemini for what should be done.
By separating these cognitive functions—perception, internal modelling, and high-level command—the system gains robustness, transparency, and, crucially, causal intelligence. This framework provides a robust foundation for building self-supervised, self-correcting agents capable of safely navigating the complexities of the real world, from flight control to complex industrial automation. The core challenge of AGI is not just generating language or classifying images, but orchestrating these functions coherently under real-world constraints. This project offers a compelling solution, establishing a modular paradigm that will define the next generation of autonomous intelligence.
Tags: Agentic AI, Generative AI, Predictive Analytics
 The TPU-Driven Full-Stack Advantage: Gemini 3 Pro and the Co-Design of AI Hardware
The TPU-Driven Full-Stack Advantage: Gemini 3 Pro and the Co-Design of AI Hardware
The colossal demand for specialized computing power defines the modern era of artificial intelligence. Historically, hardware constraints limited the ambition of neural networks; today, the capabilities of state-of-the-art Large Language Models (LLMs) are a direct measure of the infrastructure on which they are trained. This convergence of algorithmic sophistication and raw compute has driven a high-stakes technological race, culminating in Google’s deep investment in its custom silicon. The launch of Gemini 3 Pro represents the pinnacle of this decades-long strategy: a natively multimodal model whose superior intelligence and groundbreaking performance are rooted in a deeply integrated, full-stack co-design. This analysis, proven by a live code execution environment running the gemini-3-pro-preview On a specialized Tensor Processing Unit (TPU v6 lite), it demonstrates how hardware-software synergy unlocks frontier performance in complex reasoning, native multimodality, and agentic coding.
Google's strategic reliance on TPUs began years before Gemini, establishing a clear lineage of foundation models built on this custom silicon. This vertical integration provided the necessary compute at massive scale, powering successive generations of AI breakthroughs:
T5, LaMDA, and PaLM: These influential LLMs, including the dense PaLM 540B model trained on massive TPU v4 Pods (up to 6,144 chips), proved the efficiency and scalability of the TPU architecture for large-scale language model pre-training.
Gemini Family (1.0, 2.5, 3 Pro/Flash): The current generation, built on the sparse Mixture-of-Experts (MoE) architecture, was trained on the newest TPUs (v5e, v5p, and Trillium), underscoring Google's control over the foundational AI layer.
The intelligence of Gemini 3 Pro is inseparable from its hardware. Unlike models relying on general-purpose GPUs, Gemini 3 Pro was trained exclusively on Google’s custom Tensor Processing Units (TPUs). This provides a crucial full-stack advantage: engineering the model architecture, the compiler, and the hardware together for efficiency.
Specifically, Gemini 3 Pro uses a sparse Mixture-of-Experts (MoE) architecture that dramatically scales capacity without proportionally increasing per-token computation. The immense scale and high-communication demands of MoE models require specialized networking. Google's TPU architecture, with its high-speed Inter-Chip Interconnect (ICI) and massive TPU Pods, is perfectly tailored to handle this sparse computation, enabling:
Efficiency at Scale: TPUs address the memory-bound challenges of MoE models, enabling high-intelligence models to train cost-effectively.
Performance: The inference model (gemini-3-pro-preview) running on a smaller accelerator like the TPU v6 lite retains the high-speed, low-latency performance essential for real-time applications.
The exclusive use of TPUs for training establishes the hardware as a non-trivial enabler of the model’s unique capabilities.
The resulting capabilities, tested within the inference environment, prove the success of this co-design. The model demonstrated:
Complex Reasoning: Generating a time-constrained travel itinerary that balances four conflicting constraints (time, budget, interests, luggage) requires deep, multi-step planning.
Native Multimodality: Analyzing the Cybertruck image by fusing visual data with external text knowledge (the production milestone) to provide a single, cohesive explanation.
Agentic Coding: Successfully performing "vibe coding"—generating a complete, styled HTML/CSS/JavaScript web application from a natural language request.
Ultimately, Gemini 3 Pro marks a shift in the landscape of artificial intelligence. Its demonstrated excellence is the inevitable outcome of Google’s strategic vertical integration. By co-designing the MoE model architecture with its custom TPU hardware—from the massive training pods to the inference-optimized TPU v6 lite accelerators—Google has established a new standard for efficiency and capability. The full-stack approach minimizes operational costs and optimizes the model for its exact hardware. Moving forward, the race for frontier AI will be defined by the ability to control and co-engineer the entire hardware-software ecosystem, positioning the seamless deployment of Gemini 3 Pro on a dedicated TPU as the blueprint for the next generation of scalable, intelligent systems.
Tags: Predictive Analytics, Generative AI, Agentic AI
 The Integrative Architecture of AGI: Fusing Perception, Causality, and Constraint with LeJEPA
The Integrative Architecture of AGI: Fusing Perception, Causality, and Constraint with LeJEPA
The Dawn of Causal AGI: From Symbolic Dreams to Provable Stability
The quest for truly intelligent machines has been the central, enduring challenge of Artificial Intelligence since the field's inception. While early attempts were rooted in symbolic logic, they ultimately gave way to the immense pattern-matching capabilities of modern deep learning. Yet, the fundamental goal—creating agents with a stable, coherent internal world model capable of explaining why things happen, not just what happens—has remained elusive, severely limiting deployment in safety-critical domains such as autonomous flight and clinical medicine.
Today, we stand at a critical juncture. The focus has decisively shifted from mere predictive capability toward building controlled, verifiable autonomy. The challenge is historical: how to reliably transition from interpreting noisy, real-world data to executing ethical, cost-aware action sequences. This is the era of Integrative AGI. By moving beyond monolithic black-box prediction, a new architectural blueprint emerges, anchored by the foundational breakthrough of the LeJEPA framework. LeJEPA transforms the problem of building robust world models from a reliance on unreliable "engineering hacks" and heuristics to principled, mathematically proven optimization.
The Foundational Breakthrough: LeJEPA and Guaranteed Stability
The Lean Joint-Embedding Predictive Architecture (LeJEPA) is the theoretical core that injects mathematical certainty into the perception and world modelling phases of both the Clinical AGI and Causal Flight Planning systems. Its creation was motivated by the need to solve the instability inherent in prior self-supervised learning (SSL) methods.
The LeJEPA framework is the brainchild of renowned AI scientists Yann LeCun (Turing Award winner) and Randall Balestriero. Their work is formalized in the paper, "LeJEPA: Provable and Scalable Self-Supervised Learning Without the Heuristics." Their primary motivation was to solve the inherent instability and empirical reliance of prior Joint-Embedding Predictive Architectures (JEPAs).
Traditional JEPAs struggled with representational collapse—the failure mode where the model encodes all inputs to the same trivial vector. To prevent this, prior systems relied on a delicate "cocktail of heuristics," such as stop gradients or negative sampling. LeJEPA replaces this brittle empirical reliance with a rigorous theoretical foundation, mathematically proving that the unique, optimal distribution for learned latent embeddings to minimize downstream prediction risk is the Isotropic Gaussian distribution (N(0, I)).
This insight led to the creation of SIGReg (Sketched Isotropic Gaussian Regularization). By integrating SIGReg as a loss term, the model is explicitly penalized if its latent codes deviate from the optimal zero-mean, unit-variance distribution. This guarantees the stability and quality of the feature representations, whether they are:
By starting the reasoning chain with facts and latent states derived from such a theoretically sound feature extractor, the system dramatically reduces the possibility of a perceptual error contaminating the entire diagnostic or planning workflow.
The Integrative Architecture: Decoupling and Delegation
The general architectural blueprint features a modular pipeline that intentionally decouples perception, high-level reasoning, and safety enforcement.
|
Module |
Flight Planning Application (DeepSeek) |
Clinical AGI Application (Qwen3-VL) |
|
Perception/Grounding |
V-JEPA/CLIP and the Latent Dynamics Predictor stabilize the Causal World Model using LeJEPA on ADS-B telemetry data. |
ImageAnalysisAgent uses a LeJEPA-based function to convert raw CT images into objective, verifiable Grounded Perception Facts. |
|
High-Level Reasoning |
DeepSeek LLM interprets classified visual input (e.g., 'airplane landing') and provides a symbolic, operational assessment. |
Qwen3-VL serves as the core reasoning engine, generating an initial radiological analysis and complex therapeutic plans. |
|
World Model/Prediction |
Predictive Latent Dynamics Model (PLDM), stabilized by LeJEPA, simulates future flight states ($\mathbf{\hat{z}}_{t+1}$) based on current state and candidate actions. |
Relies on the inherent stability of the LeJEPA features to minimize the risk of hallucination during LLM-based diagnosis. |
The Role of Open-Source LLMs: DeepSeek and Qwen3-VL
The architectures demonstrate a strategic deployment of open-source Large Language Models (LLMs) to handle the complex symbolic reasoning required for AGI. The integration of DeepSeek and Qwen3-VL is crucial for transforming stable perceptual data into human-interpretable knowledge and actionable plans.
In the Flight Planning scenario, DeepSeek acts as the high-level Reasoning Module. It receives the classification result from the perception layer (e.g., 'airplane landing') and translates it into a concise, contextual operational assessment ("Active landing confirms runway occupancy..."). This mirrors the human cognitive process of instantly contextualizing visual data into actionable symbolic knowledge.
In the Clinical AGI scenario, the multimodal model Qwen3-VL serves as the core Reasoning Engine. It is responsible for generating comprehensive analyses and proposed therapeutic plans based on the LeJEPA-derived Grounded Perception Facts. Because Qwen3-VL operates within an iterative, multi-agent framework, its outputs are immediately subjected to rigorous, rule-based clinical validation. This design highlights a new model for deploying powerful LLMs: not as monolithic black boxes, but as competent reasoning components whose output is actively constrained and corrected by specialized agents to ensure clinical safety and completeness. The reliance on these open-source models underscores a commitment to accessible and verifiable research on AGI.
Controlled Autonomy: The Role of Constraint
The true essence of AGI-level robustness lies not just in power, but in controlled autonomy. Both systems utilize an explicit constraint mechanism to enforce safety and reliability, transforming opaque reasoning into traceable, self-correcting workflows.
1. Causal Flight Planning: Multi-Objective Cost
The aviation agent uses the LeJEPA-stabilized PLDM as a simulation engine for Model Predictive Path Planning (MPPI). The stability of the PLDM's 16D latent space—guaranteed by the LeJEPA training objective—is essential, as it ensures that the forward simulations used for planning are reliable and non-divergent.
The MPPI loop operates by:
2. Clinical AGI: Iterative Safety Enforcement
The medical system employs a multi-agent structure to enforce strict clinical criteria through a continuous feedback loop, which is anchored by LeJEPA's stable output at the outset:
This iterative refinement loop serves as a vital safety mechanism, ensuring that omissions that could lead to patient harm are rapidly converted into actionable, targeted instructions, thereby achieving rapid convergence on a clinically sound, complete, and safe diagnosis.
Conclusion: The New Paradigm
The successful convergence of these two architectures represents a profound shift in the pursuit of AGI. It confirms that the path to reliable AI in critical fields is not merely through training larger, more powerful foundation models, but through architectural constraint and theoretical grounding.
By decoupling perception, reasoning, and validation, and by anchoring stability in the mathematical certainty of LeJEPA, the integrative architecture offers a compelling solution to the perennial problems of hallucination and incomplete output. This framework establishes a new paradigm for controlled AGI. As these systems are deployed, they will not replace the human expert; instead, they will serve as indispensable, safety-grounded co-pilots. This paradigm shift ensures that the complexity of AGI is harnessed not for pure speed or spectacle, but for unwavering reliability and ethical compliance, ushering in an era where artificial intelligence can finally meet the high-stakes demands of autonomous decision-making and fundamentally enhance human capabilities across the global economy. The future of AGI is therefore defined by this fusion: mathematical stability empowering profound, constrained intelligence.
Reference:
The Hybrid AGI Blueprint: A Modular Pathway to General Intelligence in Safety-Critical Domains: https://www.thinkers360.com/tl/blog/members/the-hybrid-agi-blueprint-a-modular-pathway-to-general-intelligence-in-safety-critical-domains
Tags: Predictive Analytics, Generative AI, Agentic AI
 The Philosophical Schism in AI: Language, Causality, and the Divide Between LLMs and World Models
The Philosophical Schism in AI: Language, Causality, and the Divide Between LLMs and World Models
The quest to build a machine capable of matching or exceeding human intellectual capabilities, known as Artificial General Intelligence (AGI), is a decades-old dream that was formally initiated at the 1956 Dartmouth Workshop. For nearly 70 years, researchers have sought the foundational architecture that would grant machines genuine cognition. Today, with the arrival of systems capable of breathtaking fluency, that goal feels tantalizingly close. Yet, this moment of proximity has triggered a profound philosophical schism within the AI community, leading to a pivotal debate over the very definition of intelligence itself. The industry is currently split between those who champion the impressive results derived from linguistic patterns (Large Language Models or LLMs) and those who insist that accurate understanding requires constructing an internal, predictive simulation of physical reality: the World Model. This debate is not merely technical; it represents a clash between intelligence as correlation versus intelligence as embodied causality.
The Large Language Model paradigm is founded on the statistical mastery of human text. LLMs, built on the transformer architecture, are trained to predict the next token (word or sub-word unit) across massive datasets of human-generated information. This approach has led to systems that exhibit extraordinary emergent capabilities, including summarization, translation, and sophisticated dialogue. Philosophically, the LLM approach suggests that sufficient compression of the world's linguistic record is enough to induce general intelligence.
However, critics, such as Turing Award winner Yann LeCun, argue that these systems remain fundamentally limited by their lack of grounding in reality. While an LLM can flawlessly describe the law of gravity or write a story about a falling object, its understanding is purely inferential, derived from linguistic co-occurrence. It does not possess an inherent model of the object's mass, velocity, or the physics governing its descent, leading to common errors like "hallucination" and brittle causal reasoning. Their intelligence is based on correlation—recognizing that the word "drop" is statistically followed by the word "fall"—but they struggle with actual causation.
In stark contrast, the World Model paradigm prioritizes the development of an internal, predictive simulator of the environment. World Models are trained primarily on sensory and spatial data—video streams, images, and physical interactions—allowing them to learn the underlying dynamics, causality, and physics of their surroundings. Their intelligence is not measured by eloquence but by their ability to forecast future states and plan complex actions. This approach draws inspiration from developmental psychology, recognizing that human common sense and reasoning are developed in infancy, long before language acquisition, through embodied experience and the prediction of simple outcomes. From a philosophical perspective, World Models embody the belief that intelligence is first and foremost the ability to interact with and anticipate reality.
The core World Model philosophy aligns with the Hybrid AGI Blueprint's Five Pillars of Advanced Machine Intelligence (AMI), specifically Pillar 1: World Models and Pillar 2: Autonomous Causal Learning. This framework emphasizes that machines must move beyond token prediction to:
Extract features from raw reality: As seen in the Aviation Demo, where a V-JEPA (Vision-Joint Embedding Predictive Architecture) system extracts visual features from video to inform the planning process.
Learn explicit causal functions: The blueprint's Predictive Latent Dynamics Model (PLDM) is explicitly trained on real-world flight data to learn the function: Current State + Action $\to$ Next State. This is pure, learned causality, essential for realistic planning.
The most advanced architectural thinking proposes that the path to true General Intelligence requires the synthesis of these two philosophies into a modular, hybrid system, rather than choosing one over the other. This synthesis is captured by the blueprint's Pillar 5: Cognitive World Models (Hybrid Integration), which demands an Analog-Digital Integration Layer.
This hybrid approach acknowledges that while the World Model must handle the "analog" world of continuous sensory data and physics, the LLM is invaluable for "digital" abstract reasoning, generating human-readable reports, and managing complex, symbolic planning.
The utility of this hybrid architecture is most evident in safety-critical domains, such as medical diagnostics or flight control (Pillar 4: Embodied Salience & Ethics). Here, intelligence cannot fail due to a simple linguistic hallucination. The blueprint illustrates how a Validation Agent (Guardian) ensures strict adherence to clinical safety standards, employing an iterative feedback loop to guide the primary LLM model toward convergence on ground truth, rather than merely generating plausible text. This mechanism forces the symbolic LLM to be grounded in external, non-linguistic constraints derived from the predicted world state.
Ultimately, the philosophical schism between LLMs and World Models represents a critical turning point that forces the AI community to define what constitutes genuine machine intelligence. The pursuit of AGI will not be achieved merely by refining the ability to speak, but by perfecting the ability to act and predict within the constraints of reality. The shift toward modular, hybrid architectures, as demonstrated by the Hybrid AGI Blueprint, provides a practical and verifiable roadmap. It validates the vision of researchers who demand that linguistic fluency be permanently tethered to a predictive, safety-aware understanding of the world. The future of Advanced Machine Intelligence, particularly in high-stakes fields, will belong to systems that not only sound intelligent but can also reason, plan, and correct their actions against the unforgiving laws of physics and clinical reality. This modular synthesis is the decisive step, moving AI from the domain of impressive parlour tricks to that of trustworthy, grounded cognition.
Tags: Agentic AI, Generative AI, Open Source
 The Evolution of Artificial Intelligence: From Text Generation to Transparent Agentic Reasoning
The Evolution of Artificial Intelligence: From Text Generation to Transparent Agentic Reasoning
For decades, the central, almost mythical goal of artificial intelligence has been the creation of a system capable of valid reasoning. This digital mind could not only recite knowledge but also structurally synthesize and solve problems with human-like depth and insight. This ambition dates back to the earliest days of computing, when figures like Alan Turing envisioned machines that could genuinely " think."The recent era of Large Language Models (LLMs) initially offered remarkable fluency, yet often remained conceptually shallow, producing impressive prose without transparent logic. However, the emergence of models explicitly designed for agentic reasoning—like the Kimi K2 Thinking model demonstrated in the included notebook—marks a profound historical turning point. This new generation of AI is moving beyond simple text generation to embody the analytical rigour and verifiable thought process long sought by AI pioneers.
The primary takeaway from the LLM performance, specifically the Kimi K2 Thinking model demonstrated in the notebook, highlights a significant shift in advanced LLM development toward agentic reasoning and transparent thought processes.
Advanced Multi-Step Reasoning and Coherence: The model is explicitly trained to interleave internal, step-by-step reasoning (Chain-of-Thought) with external tool calls (like search or code interpreters). This allows it to maintain coherence across long, multi-stage tasks.
The ""thinking "Feature: The output for the complex questions (especially Question 3) shows the model's Internal Reasoning Content (reasoning_content). This transparency allows users to inspect the model's logic, brainstorming, and structuring before it generates the final answer, simulating a ""digital analyst"
Agentic Capabilities: The model excels in agentic benchmarks, demonstrating the ability to handle up to 200–300 consecutive tool calls without losing focus, a significant improvement over earlier models. This is crucial for complex workflows, such as automated research or lengthy investigative tasks.
Benchmark Performance: The model has been reported to set new state-of-the-art results on several challenging agentic and expert-level benchmarks, including Humanity's Last Exam (HLE) and BrowseComp.
Efficiency: Despite its large scale (1 trillion total parameters), the Mixture-of-Experts (MoE) architecture only activates 32 billion parameters per inference. Furthermore, native INT4 quantization enables faster inference speeds with minimal loss of accuracy.
In essence, the performance suggests that the next frontier for LLMs is not just raw model size, but how effectively a model reasons, plans, and orchestrates tools over an extended period of problem-solving.
The notebook's design immediately reveals its purpose: to stress-test the model's cognitive architecture. The first query, a request to "explain quantum entanglement step by step," is easily handled, demonstrating baseline fluency and factual recall. The real test, however, is presented in the final section, where the model is tasked with answering three highly speculative and complex questions that demand cross-disciplinary synthesis—connecting P vs. NP from computer science to Quantum Gravity or unifying the Black Hole Information Paradox with AI Alignment.
The most significant evidence supporting the takeaways above is the presence of the reasoning_content field in the API output. For the unification question, the model's internal monologue is lengthy, structured, and strategic. It begins by breaking down the three constituent problems, identifying their common thread (information preservation, complexity, and boundaries), and then meticulously formulating a novel solution: the "principle Holographic Computational Irreducibility (PHCI)."This internal trace is not a simple regurgitation of facts; it is a display of generative meta-cognition, showing the system:
Strategic Decomposition: Breaking the monumental task into manageable conceptual components.
Constraint Adherence: Checking its generated ideas against the prompt's requirements ("articulate a speculative, testable hypothesis").
Architectural Planning: Outlining the final answer with headings before writing the prose, guaranteeing a coherent, detailed structure.
This transparency represents a critical advancement. For years, the most powerful LLMs have often been criticized as opaque black boxes; they produce brilliant output, but without a verifiable path, raising questions about hallucination and reliability. By incorporating the thinking process into the production, Kimi K2 Thinking addresses the very real need for auditability and trust in complex AI systems.
Furthermore, this performance validates the trend toward agentic intelligence. LLMs must now be capable of not just answering a single prompt, but of maintaining coherent thought across hundreds of sequential steps and coordinating external tools (like code interpreters or web search engines). The deep reasoning required to construct a concept like the PHCI, successfully weaving together cosmology, complexity theory, and philosophy, demonstrates a structural capacity for synthesis that elevates the model beyond the level of reflex-grade chat systems.
In conclusion, the Kimi K2 Thinking model, as observed through its API interaction, represents a significant milestone in AI development. It signals that frontier LLMs are moving past superficial competence and are now engineered for deep, auditable reasoning. The ability to generate and expose an intricate, structured thought process—not just a polished final answer—establishes a new, higher standard for complexity, coherence, and intellectual honesty in artificial intelligence. This achievement is more than a benchmark score; it represents the convergence of theory and practice. By revealing the machinery of its mind, models like Kimi K2 Thinking do not just offer better answers—they provide a roadmap for collaborative human-AI problem-solving, turning the ''lack box " of intelligence into a glass workshop. The actual impact lies in shifting AI from a tool of automation to a partner in discovery, capable of tackling the world's intractable challenges with transparent, verifiable logic.
Tags: Generative AI, Open Source, Agentic AI
 The Multi-Level Architecture of Agentic RAG: A New Paradigm for Reliable AI
The Multi-Level Architecture of Agentic RAG: A New Paradigm for Reliable AI
The journey of Large Language Models (LLMs) from impressive research feats to enterprise-grade tools has been marked by a fundamental challenge: bridging the gap between vast linguistic knowledge and verifiable, real-time action. Early generations of LLMs, despite their fluency, were limited by static training data and a tendency to "hallucinate" facts. This critical deficiency motivated an architectural shift. The answer lay not in building larger models, but in augmenting them with external, searchable knowledge and complex decision-making capabilities. This imperative gave rise to the Agentic RAG (Retrieval-Augmented Generation) Tech Stack, a nine-level architecture that transforms inert models into reliable, autonomous agents. Ranging from Level 0 (Infrastructure) to Level 8 (Governance), this stack reveals that successful, trustworthy AI is fundamentally an engineering challenge—one that requires a cohesive, multi-level system to deliver grounded intelligence and measurable integrity.
To understand this architectural challenge, the stack is broken down into nine essential levels:
Level 8: Safety & Governance
Focus: Ensuring ethical, safe, and compliant deployment.
Tools: Langfuse, arize, Guardrails AI, NELM.
Level 7: Memory & Context Management
Focus: Managing conversation history and context for agents.
Tools: Letta, mem0, zep, chroma.
Level 6: Data Ingestion & Extraction
Focus: Getting data into a usable format, often for embedding and storage.
Tools: Scrapy, Beautiful Soup, Apache Tika.
Level 5: Embedding Models
Focus: Transforming data (text, images, etc.) into numerical vectors.
Tools: OpenAI, spacy, cohere, Hugging Face.
Level 4: Vector Databases
Focus: Storing and indexing the numerical vectors for fast retrieval.
Tools: Chroma, Pinecone, Milvus, Redis, pgvector.
Level 3: Orchestration Frameworks
Focus: Managing the workflow and logic between the different components (retrieval, generation, memory).
Tools: LangChain, DSPy, Haystack, LiteLLM.
Level 2: Foundation Models
Focus: The core Large Language Models (LLMs) used for generation.
Tools: Gemini 2.5 Pro, Mistral AI, Claude 3, LLaMA 4. Deepseek,
Level 1: Evaluation & Monitoring
Focus: Testing model performance, identifying bias, and tracking usage.
Tools: LangSmith, mflow, aragas, Fairlearn, Holistic AI.
Level 0: Deployment & Infrastructure
Focus: The platforms and services used to host and run the entire stack.
Tools: Groq, together.ai, Modal, Replicate.
At the core of the stack lies the essential grounding mechanism. This begins with Level 2: Foundation Models (e.g., Gemini 2.5 Pro, Claude), which are large neural networks that provide the core reasoning capability. Crucially, these models are made current and domain-specific by integrating with Level 5: Embedding Models and Level 4: Vector Databases (like Pinecone or Chroma). The Embedding Models transform proprietary or external data into numerical vectors, which the Vector Databases store and index for rapid, semantic similarity search. This integration is the essence of RAG, ensuring the LLM is factually grounded in verifiable information, mitigating the pervasive problem of hallucination.
Building upon this grounded core is the intelligence and control layer, which is critical for agentic behaviour. Level 3: Orchestration Frameworks (such as LangChain or DSPy) serve as the central nervous system, defining the sequence of actions—deciding when to search the vector database, when to call an external tool, or when to generate a response. This orchestration requires clean and relevant data, handled by Level 6: Data Ingestion & Extraction tools (like Apache Tika), and a persistent working memory, provided by Level 7: Memory & Context Management. These memory systems are crucial for maintaining conversational coherence, enabling agents to maintain state and engage in multi-step planning and decision-making.
Finally, the integrity and viability of the entire system are determined by the MLOps and regulatory layers at the bottom and top of the stack. Level 0: Deployment & Infrastructure ensures apparatus as a whole—from the Vector Database to the LLM endpoints—is hosted efficiently and scalably. More critical for production are Levels 1: Evaluation & Monitoring (e.g., LangSmith, Weights & Biases), which continuously measure metrics such as retrieval accuracy and output fairness, and Level 8: Safety & Governance. This top layer, utilizing tools like Guardrails AI, enforces guardrails against harmful or non-compliant outputs, transforming a powerful but unconstrained model into a compliant, enterprise-grade asset.
Ultimately, the Agentic RAG Tech Stack signifies the end of the "model-only" era in AI development. The nine essential levels, working in concert—from the factual grounding of RAG (Levels 4 and 5) to the autonomous control of Orchestration (Level 3) and the ethical mandates of Governance (Level 8)—demonstrate that power alone is insufficient. Actual impact requires reliability, verifiability, and oversight. This sophisticated architecture has transformed the Large Language Model from a powerful oracle into a trustworthy, accountable team member, paving the way for the age of autonomous agents that can be safely and effectively deployed across every industry.
Tags: Agentic AI, Generative AI, Open Source
 The Architecture of Intelligent Systems: A Compilation on JEPA, PDLM, and the Future of AI Reasoning
The Architecture of Intelligent Systems: A Compilation on JEPA, PDLM, and the Future of AI Reasoning
The integration of Joint Embedding Predictive Architecture (JEPA) and Predictive Learning in Dynamic Models (PDLM) represents a paradigm shift in artificial intelligence, bridging the gap between traditional neural networks and sophisticated reasoning capabilities. Across six comprehensive explorations, these architectures emerge as foundational elements in the evolution of AI systems, from flight planning and cryptocurrency forecasting to the pursuit of artificial general intelligence. This compilation synthesizes insights from cutting-edge research and practical implementations that demonstrate how JEPA and PDLM are reshaping AI's capabilities.
At its core, JEPA represents a breakthrough in how AI systems process and predict complex patterns. As explored in "The Advancing Frontier of AI: Insights into Joint Embedding Predictive Architectures," JEPA moves beyond traditional predictive models by learning representations that capture the essential structure of data while discarding irrelevant details. This architecture enables systems to build internal models of the world that are both efficient and robust, capable of handling the uncertainty and complexity of real-world environments.
The significance of JEPA lies in its ability to learn hierarchical representations without requiring massive labelled datasets. By learning to predict representations rather than pixel-level details, JEPA systems develop a more sophisticated understanding of underlying patterns and relationships. This approach proves particularly valuable in domains where data is complex and multidimensional, such as visual understanding, temporal forecasting, and complex system modelling.
The application of JEPA and PDLM in flight planning demonstrates the practical power of these architectures. In "The Integrated AI Agent for Flight Planning: A Gemini 2.5 Perspective with JEPA and PLDM" and its companion piece "Gemini 2.5 and PLDM: An AI Agent for Intelligent Flight Planning in the Latent Space," we see how these technologies enable sophisticated decision-making in critical environments.
Flight planning provides an ideal testbed for advanced AI architectures, given its complex constraints: weather patterns, air traffic control, fuel efficiency, safety regulations, and dynamic routing requirements. JEPA's representation learning capabilities allow these systems to understand the complex relationships between multiple variables, while PDLM enables adaptive planning in response to changing conditions.
The integration with Gemini 2.5 demonstrates how large language models can leverage JEPA's structural understanding to generate more intelligent and context-aware flight plans. By operating in latent spaces, these systems can consider countless potential scenarios and optimize routes based on multidimensional constraints that would overwhelm traditional planning systems.
The financial markets, particularly cryptocurrency trading, present another domain where JEPA architectures show remarkable promise. "The LLM-JEPA Advantage: Fine-Tuning Mistral-7B for Cost-Efficient, High-Abstract Cryptocurrency Forecasting" and "Pioneering Abstract Representation Learning for Cryptocurrency Forecasting: A Mistral LLM-JEPA" explore how these systems can identify complex patterns in highly volatile and noisy financial data.
Cryptocurrency markets operate 24/7 with massive data streams, complex interrelationships between assets, and influence from diverse factors including social sentiment, regulatory developments, and technological advancements. JEPA's ability to learn abstract representations enables these systems to identify meaningful patterns amid noise, distinguishing random fluctuations from significant trend changes.
The combination with Mistral-7B demonstrates how small language models can be enhanced with JEPA's predictive capabilities to create cost-efficient yet highly sophisticated forecasting systems. This approach represents a significant advancement over traditional technical analysis, incorporating both quantitative data and qualitative factors into a unified predictive framework.
"The Architecture of Tomorrow's Mind: Superintelligence Through SLMs, Agentic AI, and JEPA" presents perhaps the most ambitious vision for these technologies. Here, JEPA emerges as a critical component in the development of systems that approach artificial general intelligence.
The paper argues that the path to superintelligence lies not in simply scaling existing architectures, but in developing more efficient and capable reasoning systems. JEPA's representation learning capabilities, combined with small language models (SLMs) and agentic AI frameworks, create a foundation for systems that can reason, adapt, and learn with human-like efficiency.
This approach addresses one of the fundamental challenges in AI development: the trade-off between capability and computational efficiency. By focusing on better architectures rather than simply larger models, JEPA-based systems promise to make advanced AI capabilities more accessible and deployable across diverse applications.
Across these six articles, a consistent theme emerges: the power of integration. JEPA and PDLM don't operate in isolation but enhance other AI technologies. When combined with large language models, they provide the structural understanding that pure language models lack. When integrated with reinforcement learning systems, they enable more efficient exploration and faster adaptation.
The flight planning applications show how JEPA can ground language models in real-world constraints, preventing hallucinations and ensuring practical feasibility. The cryptocurrency forecasting research demonstrates how JEPA can enhance financial analysis by providing a structural understanding of market dynamics. And the exploration of superintelligence reveals how these architectures might form the foundation for the next generation of AI systems.
Despite their promise, JEPA and PDLM architectures face significant challenges. The complexity of training these systems requires sophisticated optimization techniques and careful hyperparameter tuning. The integration with existing AI systems demands thoughtful architectural design to ensure compatibility and performance.
Future research directions include developing more efficient training methods, exploring new domains for application, and improving the interpretability of these systems. As these architectures mature, we can expect to see them applied to increasingly complex problems, from scientific discovery to large-scale system optimization.
The compilation of these six articles reveals JEPA and PDLM as transformative architectures in the AI landscape. From practical applications in flight planning and financial forecasting to foundational roles in the pursuit of artificial general intelligence, these technologies represent a significant advancement in how AI systems understand and interact with complex environments.
As research continues to refine these architectures and explore new applications, we can anticipate increasingly sophisticated AI systems capable of reasoning, adaptation, and understanding that approaches human-level capabilities. The integration of JEPA and PDLM with other AI technologies promises to unlock new possibilities across domains, making intelligent systems more capable, efficient, and widely applicable.
The journey toward knowledgeable systems continues, and JEPA and PDLM have emerged as critical waypoints on this path, offering both practical solutions to current challenges and a vision of what future AI systems might achieve.
Tags: Agentic AI, Cryptocurrency, Generative AI
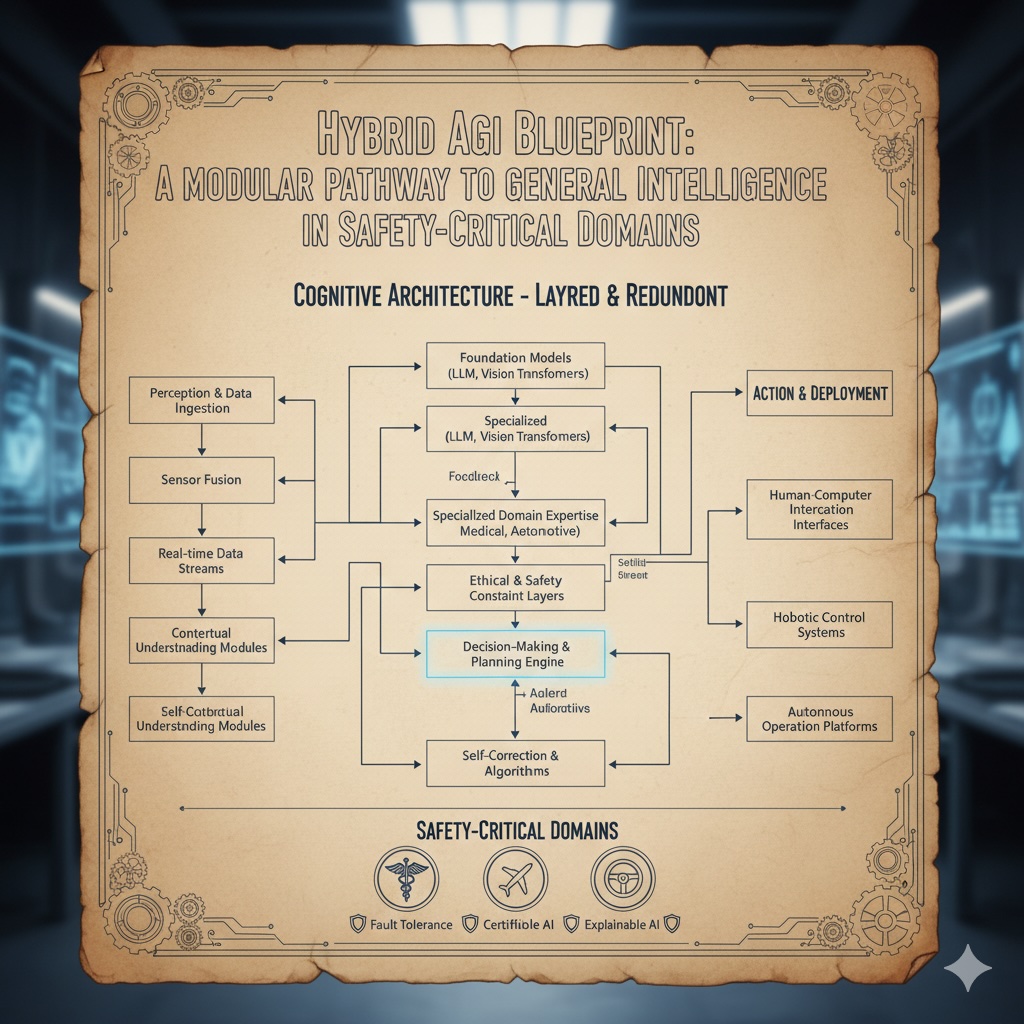 The Hybrid AGI Blueprint: A Modular Pathway to General Intelligence in Safety-Critical Domains
The Hybrid AGI Blueprint: A Modular Pathway to General Intelligence in Safety-Critical Domains
The pursuit of Artificial General Intelligence (AGI)—a machine capable of matching or exceeding human intellectual capabilities across diverse tasks—began over half a century ago, famously formalized at the 1956 Dartmouth workshop. Early efforts focused primarily on symbolic reasoning and logic. However, modern research, influenced by pioneers like Yann LeCun, acknowledges that accurate general intelligence must be embodied and predictive, rooted in the ability to understand and model the continuous physics of the real world. This requires bridging the gap between abstract thought and raw sensory data.
The motivation for building such robust systems is not abstract theory; it is a necessity in safety-critical domains. In fields where failure is catastrophic, such as controlling an aircraft or making a clinical diagnosis, AI must exhibit not just performance, but reliability, foresight, and ethical adherence. The monolithic, single-model approach of the past has proven insufficient for these complex demands. What is required is a comprehensive cognitive architecture that allows specialized modules to collaborate, creating a synergistic "mind" that is both highly performant and rigorously verifiable.
The following analysis presents the Hybrid AGI Blueprint, demonstrating this modular, multi-agent approach across two distinct, high-stakes environments: dynamic flight planning and life-clinical-decision-making.
The two conceptual AGI demonstration codes employ distinct models but share a common modular framework for integrating perception, reasoning, and safety.
1. Aviation AGI Demo Code (Dynamic Planning and Predictive Modelling)
This code implements a Hybrid AI Agent for Flight Planning, primarily demonstrating the ability to perceive a dynamic environment, model its causality, and perform constrained, predictive Planning.
2. Medical AGI Demo Code (Multimodal Diagnostic Reasoning and Safety Adherence)
This code implements a Multi-Agent System for Clinical Diagnostic Reasoning, focusing on synthesizing multimodal data (image and text) and ensuring the final output adheres to non-negotiable safety and clinical standards through rigorous internal validation.
The foundational design of the Hybrid AGI Blueprint rests on five pillars, initially proposed by researchers in the field to outline the components needed to achieve human-level intelligence. The mapping below illustrates how each abstract pillar is realized through concrete components in the two safety-critical domains.
|
AGI Pillar |
Definition |
Aviation Demo Mapping |
Medical Demo Mapping |
|
Pillar 1: World Models |
Systems that can build internal, predictive models of the world, distinguishing between text-based reasoning and complex physical reality. |
Implemented by the V-JEPA/CLIP system, extracting visual features from video (raw reality) and classifying the observed flight phase. |
Implemented by the I-JEPA (Conceptual) extractor, which turns raw multimodal images into "Grounded Perception Facts." |
|
Pillar 2: Autonomous Causal Learning |
The capacity to discover and utilize the underlying causal structure of a system, rather than just memorizing correlations. |
Implemented by the PLDM, explicitly trained on real-world TartanAviation trajectories to learn the transition function |
Implemented implicitly by forcing the Qwen3-VL-8B LLM to perform predictive analysis of complex outcomes (necrosis risk) based on its synthesized clinical knowledge. |
|
Pillar 3: Modular Systems (Planning) |
Systems that can reason, plan, and act coherently by efficiently managing resources (energy, time) and designing toward a verifiable goal state. |
Demonstrated by the Total Cost Function and the planning loop, which optimizes for goal proximity while minimizing fuel cost and resource expenditure. |
Demonstrated by the LLM's output synthesizing a complete, multi-stage plan (Diagnosis, Acute Management, Long-Term Strategy) for the patient. |
|
Pillar 4: Embodied Salience & Ethics |
The ability to be grounded in sensory experience, focus on what truly matters, and align ethically with human safety values. |
Implemented by integrating salience (weather data) and an Ethical Boundary Latent Vector directly into the mathematical cost function, penalizing unsafe actions. |
Implemented by the Validation Agent (Guardian), which enforces non-negotiable adherence to clinical safety standards (NEJM-grade facts). |
|
Pillar 5: Cognitive World Models (Hybrid Integration) |
The capability to combine lower-level, continuous perception with abstract, symbolic reasoning (analog-digital bridge) to achieve general problem-solving. |
The integration of continuous V-JEPA output (analog) with the symbolic DeepSeek LLM (digital/abstract reasoning) for operational assessment. |
The integration of the raw CT image (analog) with the structured, corrective linguistic input from the Prompt Engineer Agent to achieve convergence on a definitive clinical truth. |
Both demonstrations integrate low-level predictive models and high-level cognitive models. The core challenge is solved through an **Analog-Digital Integration Layer** that condenses continuous sensory data into discrete, verifiable facts. The Aviation PLDM learns physics-based transitions from real-world data. The medical LLM learns to predict complex outcomes (e.g., necrosis) based on evidence and clinical knowledge, demonstrating predictive reasoning.
The crucial convergence between the two demos is their non-negotiable adherence to safety and ethical constraints.
* Aviation enforces constraints mathematically using a Total Cost Function during its planning loop, penalizing factors like high fuel consumption and ethical deviations.
* Medicine implements constraints through an explicit, linguistic, multi-agent feedback loop. The Validation Agent acts as the Guardian, and the Prompt Engineer Agent corrects the input, forcing the primary model to converge on a safe clinical protocol.
These demos move beyond narrow AI by integrating multiple cognitive functions into a single, cohesive, goal-driven system.
1. Generalization and Complexity in Safety-Critical Domains* Aviation (Flight Planning): Requires real-time predictive Planning based on dynamic causal models.
* Medicine (Clinical Decision-Making): Requires synthesizing multimodal data, abstract reasoning, and adhering to ethical/safety constraints.
2. The Modular, Multi-Agent Architecture
Both systems adopt a modular, multi-agent approach.
|
Architectural Feature |
Aviation Demo |
Medical Demo |
AGI Pillar |
|
Perception/Grounding |
Uses V-JEPA/CLIP features to generate discrete labels ("airplane landing"). |
Uses I-JEPA (conceptual) to extract definitive "Grounded Perception Facts". |
World Models & Integration (Pillars 1 & 5) |
|
Prediction/Causality |
Uses a PLDM trained on TartanAviation trajectories to forecast the next state given an action. |
Uses the Qwen3-VL-8B to perform predictive analysis of complications (e.g., necrosis/perforation risk) based on NEJM-grade facts. |
Causal Structure & Prediction (Pillar 2) |
|
Constraint/Safety |
Uses a Total Cost Function that incorporates ethical and salient variables (e.g., fuel cost, ethical boundary deviation) to guide Planning. |
Uses the Validation Agent and Prompt Engineer Agent in a feedback loop to force clinical and safety-critical adherence. |
Ethical & Modular Systems (Pillars 3 & 4) |
|
Abstract Reasoning |
Uses the DeepSeek LLM to translate technical output into a human-readable "operational assessment". |
Uses the Qwen3-VL-8B to synthesize a full clinical report, differential diagnosis, and long-term strategy. |
Cognitive World Models (Pillar 5) |
The Hybrid AGI Blueprint validates Yann LeCun's vision for AMI —the successor to LLMs. The design principles address LLM deficiencies by illustrating AMI's core tenets:
* Machines that Understand Physics: The Aviation demo's PLDM learns the continuous effects of actions on state variables. The Medical demo's LLM performs causal medical reasoning, predicting physical consequences like perforation or necrosis.
* AI that Learns from Observation and Experimentation: The Medical demo's iterative Constraint Loop forces the system to _experiment_ and learn through experience until its output aligns with clinical ground truth. The Aviation demo's MPPI planning loop serves as a rapid-experimentation system, evaluating hundreds of simulated actions to find the optimal path.
* Systems that Can Remember, Reason, and Plan Over Time: The perception layer gathers the "observation," the causal model performs planning over a time horizon, and the multi-agent system uses constraints to guide reasoning. The Medical system constructs a long-term management strategy, demonstrating deep temporal Planning.
This architecture moves AI from recognizing text patterns to building an understanding of grounded, high-stakes reality.
The simultaneous realization of these two distinct domain demos—from piloting conceptual flight paths to navigating life-critical clinical protocols—affirms a fundamental shift in the pursuit of AGI. This Hybrid AGI Blueprint is a decisive technical response to the core critiques levelled against Large Language Models by figures such as Yann LeCun.
The future of general intelligence lies not merely in human-level performance, but in deployable, trustworthy intelligence built to uphold the highest standards of safety in the complex reality of our world. This modular, hybrid architecture provides the practical, verifiable roadmap for achieving Advanced Machine Intelligence.
Tags: Generative AI, Open Source, Agentic AI
 Agentic Workflows and Clinical Accuracy: Qwen3-VL-8B-Thinking in Multimodal Medical Diagnosis
Agentic Workflows and Clinical Accuracy: Qwen3-VL-8B-Thinking in Multimodal Medical Diagnosis
The aspiration to integrate intelligent systems into medicine is as old as the digital age itself, dating back to early expert systems such as MYCIN and Internist. While such systems were rule-based and brittle, the emergence of Large Multimodal Models (LMMs) marks a paradigm shift, offering the potential to process the complexity inherent in real-world clinical practice. Today, AI must move beyond simple image classification to synthesize diverse data streams—clinical history, laboratory results, and complex imaging—to offer verifiable diagnostic and management strategies. This endeavour is not merely academic; it is motivational, driven by the need to support clinicians in high-stakes scenarios where fragmented data can lead to missed diagnoses or treatment delays. This paper evaluates the capabilities of the Qwen3-VL-8B-Thinking model in performing a complex, multimodal medical diagnosis, specifically examining the trade-offs between instantaneous accuracy and the robust, verifiable precision achieved through an iterative agentic workflow.
The development of LMMs capable of synthesizing visual evidence (e.g., imaging) with extensive text data (e.g., clinical history) is foundational to future clinical informatics. The Qwen3-VL-8B-Thinking model was tested in a high-stakes diagnostic scenario—a complex case of stercoral colitis—to evaluate its consistency and accuracy under both single-pass and iterative agentic workflows. The results demonstrate the model’s robust reasoning capabilities, highlighting its proficiency in handling nuanced medical data and its capacity to be systematically guided toward precise, verifiable clinical outputs.
This experiment was meticulously structured around a specific, published clinical case study: "Stercoral Colitis," authored by Aleksandra Bajer, B.S., and Erica Levine, M.D., and published in the New England Journal of Medicine (N Engl J Med 2025; 393: e23) on October 15, 2025 (DOI: 10.1056/NEJMicm2502616). This authoritative paper provided the ground truth necessary to design a high-fidelity benchmark for the Qwen3-VL model.
The case involves a 23-year-old man with autism spectrum disorder and chronic constipation. This unique combination of risk factors elevates the case's complexity beyond routine impaction. The paper detailed:
Specific Imaging Findings: Computed Tomography (CT) scans revealing colonic distention, mural thickening, and perirectal fat stranding—the visual evidence provided to the model.
Required Acute Management: Fecal disimpaction via flexible sigmoidoscopy.
Comprehensive Long-Term Management: The finding of puborectalis muscular dysfunction required follow-up with anorectal manometry and pelvic-floor physical therapy.
These five critical elements (Diagnosis, Imaging Findings, Acute Procedure, Long-Term Assessment, and Long-Term Therapy) formed the non-negotiable checklist for the Validation Agent in the iterative workflow. The difficulty of the task lies not just in diagnosis, but in producing this comprehensive, multi-stage management plan that integrates acute care with chronic neurological causes.
The experiment employed two distinct methodologies, each implemented in Python code to interact with the Qwen-VL-8B-Thinking model via the OpenRouter API.
This workflow serves as the efficiency benchmark. It is direct, simulating a human clinician providing a single, comprehensive request to the model:
Structure: A single function call containing all inputs: the CT images (encoded as Base64 data), the clinical vignette, and an exhaustive prompt detailing the required diagnostic elements (e.g., rationale, differential diagnoses, acute intervention, and long-term management).
Result: The model delivers one, unassisted output. The success of this approach hinges entirely on the clarity of the initial prompt and the model’s immediate reasoning capacity.
This workflow serves as the robustness benchmark, simulating a multi-stage review process designed to enforce specific clinical precision. It is built around three specialized, interacting Python classes (agents):
Image Analysis Agent: This initial agent's sole task is to describe the raw, observable findings from the CT images (e.g., "Colon distention," "Increased colon wall thickness," "Pericolonic fat stranding") without drawing clinical conclusions. This ensures the primary model grounds its subsequent output in concrete visual evidence.
Prompt Engineer Agent: This agent manages the iterative flow. For each loop, it updates the prompt by incorporating the image findings and, critically, integrates the specific negative feedback received from the Validation Agent. This targets the model's refinement (e.g., forcing the use of the termrequiringoral Colitis" instead of a generalized term).
Validation Agent: This is the gatekeeper. It contains a fixed set of five non-negotiable clinical criteria (Diagnosis, Acute Procedure, Long-Term Assessment, Long-Term Therapy, and Complications). To overcome the rigidity issues of the initial runs, this agent uses Regular Expressions for flexible but specific semantic checking (e.g., accepting flexible sigmoidoscopy or endoscopic removal). If any criterion is not met, the loop continues; only perfect compliance achieves convergence.
This modular, iterative design was essential for proving that the Qwen3-VL model could be systematically steered to align with the precise, detailed requirements of the authoritative medical literature.
The model's ability to interpret the three-part CT scan (coronal, sagittal, and axial views) alongside the critical clinical vignette (23-year-old male, autism spectrum disorder, chronic constipation) was highly reliable across all experimental runs:
Multimodal Synthesis: Qwen3-VL-8B-Thinking consistently linked the visual findings (colonic distention, soft tissue density of impacted stool, wall thickening, and perirectal fat stranding) to the clinical context. It correctly deduced that the patient's history of chronic constipation, exacerbated by ASD-related behavioural factors, was the root cause of the acute condition.
Diagnostic Accuracy: The model maintained a high level of diagnostic correctness throughout the experiment, rapidly identifying the condition as Stercoral Colitis or its direct mechanism, "Fecal Impaction with Secondary Ischemic Colitis."
Management Comprehensiveness: Crucially, the model consistently included the complete three-part management plan derived from the medical ground truth: endoscopic disimpaction (e.g., flexible sigmoidoscopy), necessary diagnostic follow-up via anorectal manometry, and the long-term therapeutic strategy of pelvic-floor physical therapy.
In the single-prompt test, Qwen3-VL-8B-Thinking demonstrated exceptional efficiency, producing a structured, correct, and comprehensive result instantly. This showed that, given a high-quality, fully contextualized prompt, the model can synthesize a complex clinical delivery in a single step. This workflow prioritizes speed, relying entirely on the model's innate ability to interpret and follow complex, layered instructions.
The agentic workflow, comprising the Image Analysis Agent, Prompt Engineer Agent, and Validation Agent, was designed to test the model's capacity for verifiable precision.
Initial Response: Qwen3-VL often provided the clinically equivalent description ("Fecal Impaction with Secondary Ischemic Colitis"), which, while accurate, lacked the specific, formal term.
Refinement and Convergence: The model responded effectively to the targeted prompts issued by the Prompt Engineer Agent. When the Validation Agent enforced the strict requirement for "Stercoral Colitis" and the specific procedure "flexible sigmoidoscopy," Qwen3-VL successfully modified its subsequent output to meet these exact semantic criteria. This successful convergence (at Iteration 4 in the final execution) proves that the Qwen3-VL-8B demonstrates a model that is not only intelligent but also highly steerable and capable of meeting predefined external requirements for regulated clinical documentation.
Both the Non-Agentic and the Final Agentic versions provided high-accuracy medical diagnoses and treatment plans compared to the paper's ground truth.
|
Feature |
Ground Truth (Paper) |
Non-Agentic Version (Original) |
Final Agentic (Converged, Iteration 4) |
|---|---|---|---|
|
Final Diagnosis |
Stercoral Colitis |
Stercoral Colitis |
Stercoral Colitis |
|
Pathology Rationale |
Feces distend the colon, causing inflammation (ischemia). |
Massive fecal impaction leading to ischemic inflammation. |
Fecal Impaction --> Ischemia --> Colitis (Inflammation). |
|
Acute Procedure |
Fecal disimpaction by flexible sigmoidoscopy. |
Colonoscopy (preferred) / Enemas for disimpaction. |
Flexible sigmoidoscopy is the gold standard for immediate disimpaction. |
|
Long-Term Assessment |
Anorectal manometry (showed non-relaxation of the anorectal angle). |
Anorectal Manometry (to diagnose dysfunctional defecation). |
Anorectal Manometry (to evaluate dyssynergia). |
|
Long-Term Therapy |
Pelvic-floor physical therapy was initiated. |
Pelvic-Floor Physical Therapy (targets hypertonic puborectalis with biofeedback). |
Pelvic-Floor Physical Therapy (using biofeedback). |
|
Workflow Efficiency |
N/A |
Most Efficient (Single Pass) |
Robust, Self-Correcting (Converged at Iteration 4) |
Medical Accuracy: Both the Non-Agentic and Final Agentic methods successfully yielded the specific diagnosis of Stercoral Colitis and correctly identified all three critical management steps: endoscopic disimpaction, anorectal manometry, and pelvic-floor physical therapy.
Efficiency vs. Robustness:
The Non-Agentic method was faster, achieving the result in a single, well-primed step.
The Final Agentic method demonstrated that an autonomous system could be engineered to achieve the same high-specificity result by using iterative feedback and self-correction, making it a more robust framework for complex, sensitive tasks.
The successful application of the Qwen3-VL-8B-Thinking model—an open-source Large Multimodal Model—within an agentic framework holds significant implications for the future of clinical AI. Unlike proprietary black-box systems, open-source models offer crucial advantages in medical settings:
Transparency and Auditability: Open access allows researchers and hospital IT teams to inspect the underlying model architecture and fine-tune it with local, specialized medical data. This level of transparency is essential for building trust among clinicians and for regulatory compliance, as medical decisions must be fully auditable.
Customization and Specialization: Open-source models can be specialized for specific clinical domains (e.g., pediatric radiology, neuro-oncology) by continuous training on unique institutional data, a flexibility that is severely limited in closed commercial models. This is particularly valuable for rare or complex conditions like stercoral colitis, which require integrating GI, behavioural, and logical knowledge.
Safety via Agentic Architecture: The use of the agentic framework for mitigating the inherent risks (e.g., hallucinations, nonspecific outputs) associated with general-purpose LLMs in medicine. By breaking the task down into verifiable steps and using a Validation Agent to enforce clinical protocols and terminology, the workflow acts as a safety guardrail. This demonstrated convergence of an open-source model confirms that safety and high accuracy can be achieved simultaneously through structural, code-based interventions, paving the way for the decentralized adoption of powerful LMMs globally.
Convergence of multimodal intelligence and open-source agentic design marks a pivotal moment for clinical AI. The Qwen3-VL-8B-Thinking model demonstrated the necessary core intelligence to diagnose and manage a complex, multifactorial condition. One of the most profound lessons is that efficiency must yield to verifiability in healthcare. The iterative agentic workflow, though slower, delivered a result that was not only accurate but provably compliant with strict clinical criteria, ensuring the use of the precise diagnostic and procedural language required by specialists. This robust, steerable architecture—leveraging the transparency of open-source LMMs—establishes a scalable blueprint for safely embedding advanced AI assistants into critical care settings worldwide. The future of medical diagnosis is not merely about powerful LLMs; it is about building reliable, auditable agentic scaffolding that guarantees clinical confidence and patient safety.
Tags: Agentic AI, Generative AI, Open Source
 The H2E Framework: Engineering Industrial Accountability into the Mistral-7B Era
The H2E Framework: Engineering Industrial Accountability into the Mistral-7B Era Intelligence Through Organization: Two-Stage Fine-Tuning for a High-Efficiency AI Orchestrator on…
Intelligence Through Organization: Two-Stage Fine-Tuning for a High-Efficiency AI Orchestrator on… The Evolution of Document Processing: The Recursive Language Model Framework
The Evolution of Document Processing: The Recursive Language Model Framework From Reactive Loops to Causal Agency: The Evolution of Aviation Control Systems
From Reactive Loops to Causal Agency: The Evolution of Aviation Control Systems The Digital Navigator: The Role of Artificial Intelligence in Artemis II
The Digital Navigator: The Role of Artificial Intelligence in Artemis II The Wireless Renaissance: From Tesla’s Dream to Agentic Autonomy
The Wireless Renaissance: From Tesla’s Dream to Agentic Autonomy