Jul11

The intricate process by which a linear chain of amino acids folds into a unique, three-dimensional structure is fundamental to all biological life. This "protein folding problem" is notoriously complex, yet its understanding is crucial for advancements in medicine, biotechnology, and material science. The advent of artificial intelligence presents powerful new avenues for tackling this challenge. As demonstrated by a recent AI agent system, a modular, multi-agent approach can effectively dissect and address various facets of protein folding, from data acquisition to ethical considerations, showcasing a sophisticated framework for scientific inquiry.
At the heart of this innovative approach lies the multi-agent paradigm. Instead of a monolithic AI attempting to solve the entire problem, the system employs several specialized AI agents, each endowed with distinct expertise and a set of tools. This modularity offers significant advantages: it allows for the division of labour, promotes scalability, and enables each agent to specialize in a specific domain, thereby enhancing efficiency and accuracy. This specialization reflects the collaborative nature of real-world scientific research, where experts from various fields come together to achieve a common goal, inviting you to be part of this collaborative journey.
The practical application of the MISTRAL AI system's conceptual framework is vividly illustrated through the agents' outputs. The Protein Sequence Data Agent, acting as a biological librarian, swiftly fetches an amino acid sequence and associated metadata for a given protein ID, even identifying existing experimental 3D structures. This immediate access to foundational data is a clear demonstration of the system's capabilities.
Following this, the Folding Prediction & Simulation Agent steps in, conceptually simulating the dynamic process of folding. While a short amino acid sequence might prove insufficient for a meaningful prediction, the agent can still outline the process of molecular dynamics simulation, detailing how minor structural fluctuations might occur over a short period, such as 10 nanoseconds. This highlights the agent's understanding of the underlying scientific principles, even when precise data is limited.
The code demonstrates the architecture and functionality of an AI agent system designed for protein folding analysis. The core concept is to use a multi-agent system built with the Mistral AI SDK to simulate a complex scientific workflow. The system is structured around several specialized agents, each responsible for a specific domain task:
Conceptual Simulation: The demonstration utilizes 'mock' functions to simulate the behaviour of complex scientific processes (such as AlphaFold or GROMACS), illustrating how agents would interact in a real-world scenario without requiring actual high-performance computing resources. This showcases the system's ability to handle complex scientific processes, instilling confidence in its capabilities. The overall goal is to showcase how AI agents can be configured and tested to automate a scientific workflow, explicitly addressing the challenges of protein folding and analysis.
The final output of the code, as presented in the provided code, summarizes the results of the executed test cases and the interactions between the agents. The code execution output demonstrates that the AI agents successfully performed their designated tasks using the conceptual (mock) tools defined in the notebook.
Here is a summary of the final output for each test case:
Further along the analytical pipeline, the Misfolding Analysis & Intervention Agent takes center stage. Protein misfolding is implicated in numerous diseases, making its identification paramount. This agent can pinpoint 'hotspots' – specific regions within a protein prone to misfolding or aggregation. By analyzing simulated data, it identifies areas, such as residues 600-610 and 980-990 in a hypothetical protein, attributing their propensity for misfolding to hydrophobic patches. Such insights are invaluable for understanding disease mechanisms and designing therapeutic interventions. Finally, to consolidate these disparate findings, the Result Synthesis & Interpretation Agent weaves together the predicted structures, folding dynamics, and misfolding analyses into a comprehensive report, complete with confidence scores and potential chaperone recommendations. This agent transforms raw data and analytical insights into actionable knowledge, demonstrating the power of AI in generating structured scientific summaries and empowering you with comprehensive information.
Beyond the purely scientific aspects, the system also incorporates a crucial dimension: ethical consideration. The Historical & Ethical Context Agent provides a broader perspective, capable of recalling significant milestones in protein science, such as Cyrus Levinthal's paradox, which underscored the immense complexity of protein folding.
In essence, this multi-agent AI system for protein folding exemplifies a powerful approach to tackling complex scientific problems. By breaking down a grand challenge into manageable, specialized tasks handled by interconnected agents, the system demonstrates how AI can facilitate comprehensive analysis, accelerate discovery, and even integrate ethical foresight into the scientific process. While the current demonstration utilizes conceptual mock data, the underlying framework lays a robust foundation for future AI-driven research, promising to unlock more profound insights into protein behaviour and its implications for human health.
Keywords: Agentic AI, Generative AI, Open Source
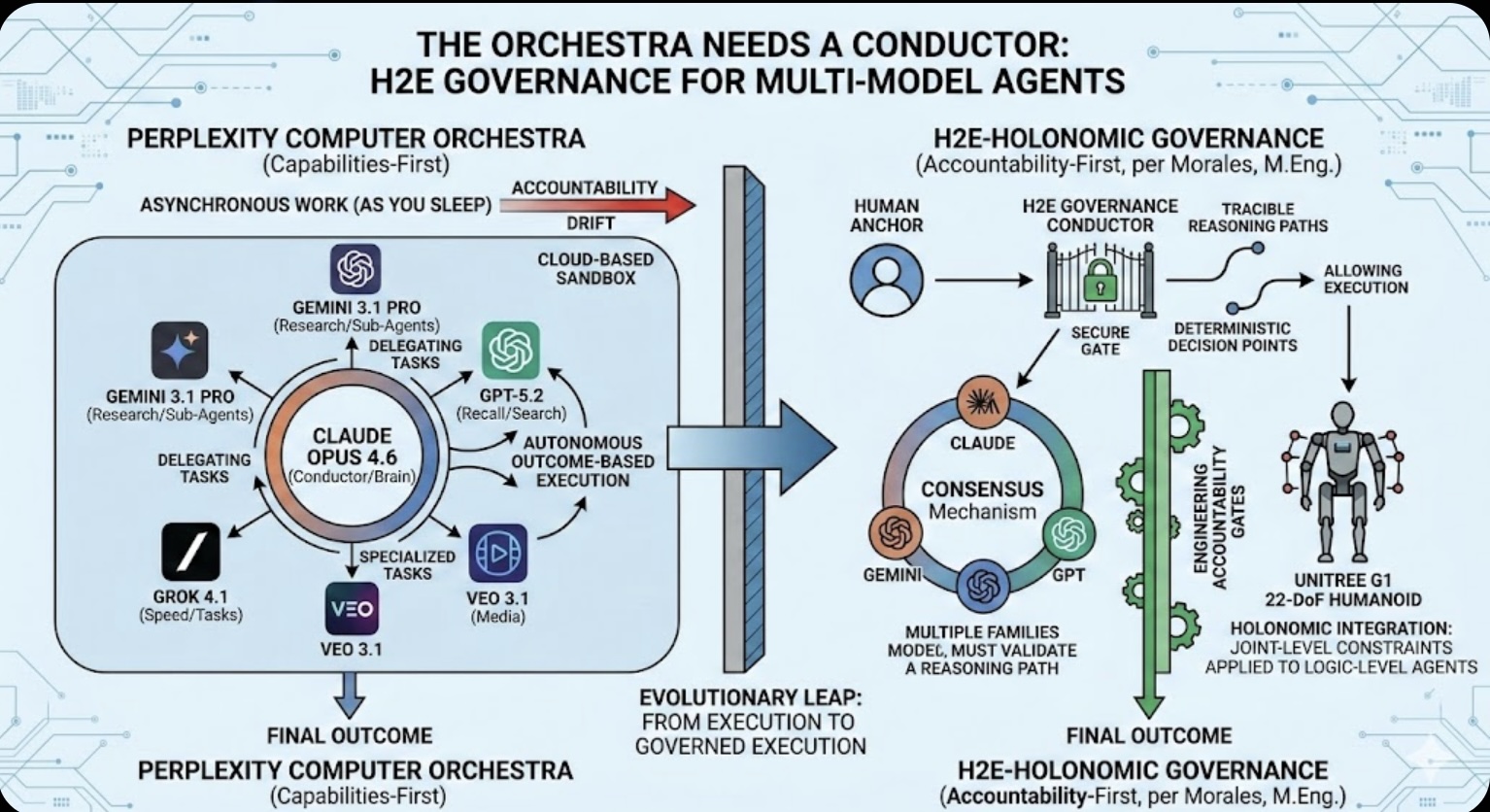 The Orchestra Needs a Conductor: Why Multi-Model Agents Require H2E Governance
The Orchestra Needs a Conductor: Why Multi-Model Agents Require H2E Governance The Role of Memory in Modern-day Business
The Role of Memory in Modern-day Business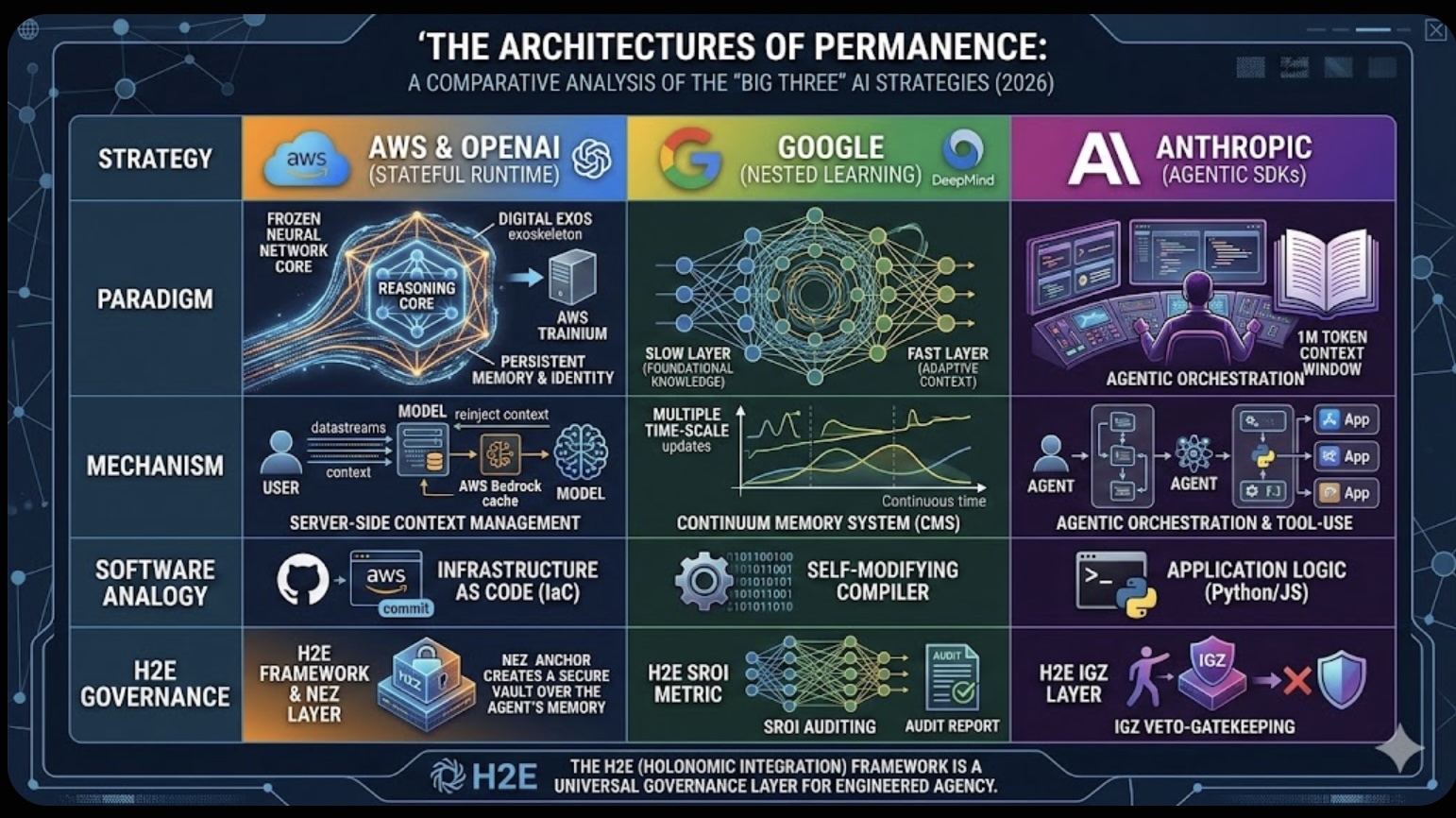 The Architectures of Permanence: A Comparative Analysis of the "Big Three" AI Strategies (2026)
The Architectures of Permanence: A Comparative Analysis of the "Big Three" AI Strategies (2026) Friday’s Change Reflection Quote - Leadership of Change - Change Leaders Enable Generational Advancement
Friday’s Change Reflection Quote - Leadership of Change - Change Leaders Enable Generational Advancement The Corix Partners Friday Reading List - February 27, 2026
The Corix Partners Friday Reading List - February 27, 2026