Jul01

The landscape of scientific inquiry is rapidly evolving, driven by the increasing complexity of grand challenges that defy traditional, single-disciplinary approaches. From the mysteries of the universe to the intricacies of life at the molecular level, these problems demand innovative solutions. A promising paradigm emerging to meet this demand is the development of modular AI agent frameworks, which leverage diverse large language models (LLMs) and specialized tools to orchestrate sophisticated problem-solving. This approach, exemplified by the MSTRAL AI Agents framework, provides a powerful blueprint for accelerating discovery, sparking curiosity, and inspiring exploration, as demonstrated by its conceptual application to the notoriously challenging protein folding problem.
The code illustrates a conceptual framework for developing and evaluating AI agents intended to address complex scientific challenges. The core idea is to break down a significant, multifaceted problem (like understanding protein folding or proving relativity) into smaller, manageable sub-problems, each handled by a specialized AI agent. Here's the breakdown of the concept:
Based on the code, two different Large Language Models (LLMs) are used for the AI agents, both developed by Mistral AI:
A crucial strategic advantage of this modular design lies in its capacity to incorporate diverse LLMs. The framework enables different agents to be powered by various underlying large language models, each selected for its specific strengths and capabilities. For instance, an agent tasked with broad knowledge retrieval, such as a "Protein Sequence Data Agent," might utilize a powerful model like mistral-large-latest. This model's "large-latest" designation suggests it is optimized for comprehensive understanding and complex reasoning across vast datasets, making it ideal for fetching diverse scientific information. Conversely, agents focused on more analytical, conceptual, or synthesis-oriented tasks, like the "Folding Prediction & Simulation Agent" or the "Result Synthesis & Interpretation Agent," might employ a "medium-latest" model. The magistral-medium-latest model noted as the primary Mistral AI model for these agents in the provided context, is likely selected for its balance of robust analytical capabilities and computational efficiency. This strategic matching of LLM capabilities to agent-specific tasks ensures that each component of the problem-solving pipeline is handled by the most suitable AI, optimizing both performance and resource utilization.
The practical utility of this framework is vividly illustrated by its conceptual application to the protein folding problem in bioscience. This challenge, encapsulated by Levinthal's Paradox, seeks to understand how proteins rapidly achieve their precise three-dimensional structures and, conversely, how misfolding leads to debilitating diseases.
The final output demonstrates the successful execution of refactored AI agents designed to tackle the protein folding problem, leveraging the Mistral AI Agents framework. The agents were successfully created and interacted with their respective mock tools, responding relevant to the bioscience field. Specifically, the output shows:
The "Protein Sequence Data Agent" successfully retrieves mock protein sequences and experimental structure data, laying the groundwork for analysis. The "Folding Prediction & Simulation Agent" conceptually attempts to predict protein structures and simulate molecular dynamics, thereby demonstrating the modelling aspect. The "Misfolding Analysis & Intervention Agent" identifies hypothetical misfolding hotspots and suggests interventions, showcasing its role in disease understanding. All these findings are then consolidated by the "Result Synthesis & Interpretation Agent" into a comprehensive report. Furthermore, the "Historical & Ethical Context Agent" offers a broader perspective, discussing milestones such as Levinthal's Paradox and analyzing the ethical implications of cutting-edge bioscience applications, including CRISPR for proteinopathies. The output demonstrates the agents' ability to process queries, invoke their specialized tools (even if mocked), and generate domain-specific responses, showcasing the framework's potential for tackling real-world scientific complexities.
The implications of such AI agent frameworks for scientific discovery are profound. By automating and intelligently orchestrating complex research workflows, these systems can accelerate hypothesis generation, data analysis, and experimental design. They offer the capacity to navigate and synthesize vast amounts of information, identify subtle patterns that human researchers might miss, and explore computational spaces far more efficiently. This represents a significant step beyond simple automation, moving towards a future where AI agents act as intelligent, collaborative partners in the scientific process, freeing human researchers to focus on higher-level conceptualization and interpretation. The modularity and adaptability of this framework suggest that its applicability extends beyond bioscience to other grand challenges, including drug discovery, materials science, climate modelling, and beyond.
In conclusion, the conceptual framework demonstrated by the Gemini 2.0 AI Agents, with its emphasis on modular AI agents, diverse LLM utilization, and specialized tool use, represents a compelling new paradigm for scientific problem-solving. By intelligently decomposing complex challenges and orchestrating specialized AI components, this approach offers a powerful pathway to unravelling some of the most enduring mysteries in science, ushering in an era of accelerated discovery and innovation.
Keywords: Agentic AI, AI, Open Source
 Lateral Moves: The Most Overlooked Succession Strategy in Companies
Lateral Moves: The Most Overlooked Succession Strategy in Companies The Asset Play: Timing, Structure & Global Arbitrage
The Asset Play: Timing, Structure & Global Arbitrage 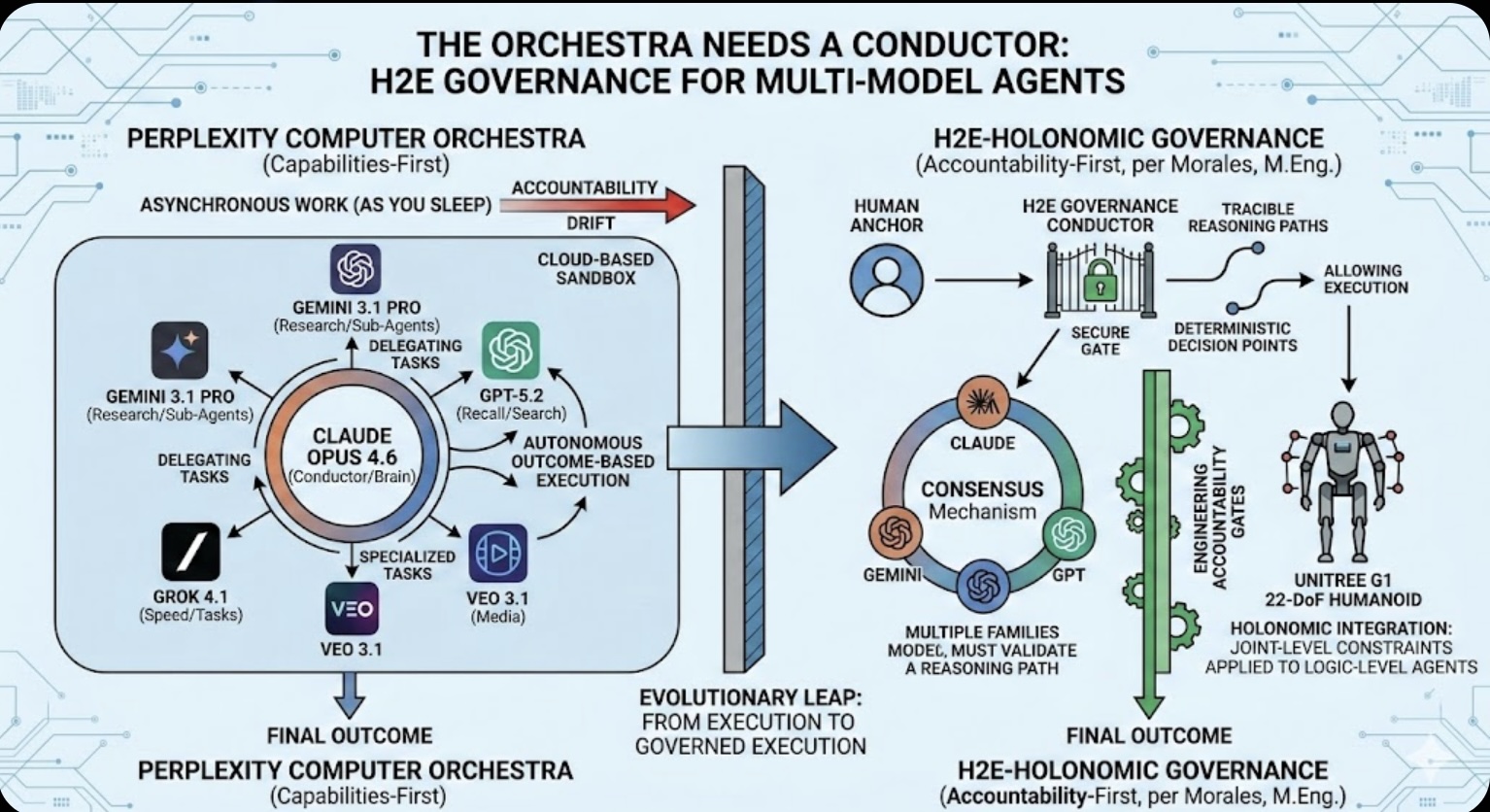 The Orchestra Needs a Conductor: Why Multi-Model Agents Require H2E Governance
The Orchestra Needs a Conductor: Why Multi-Model Agents Require H2E Governance The Role of Memory in Modern-day Business
The Role of Memory in Modern-day Business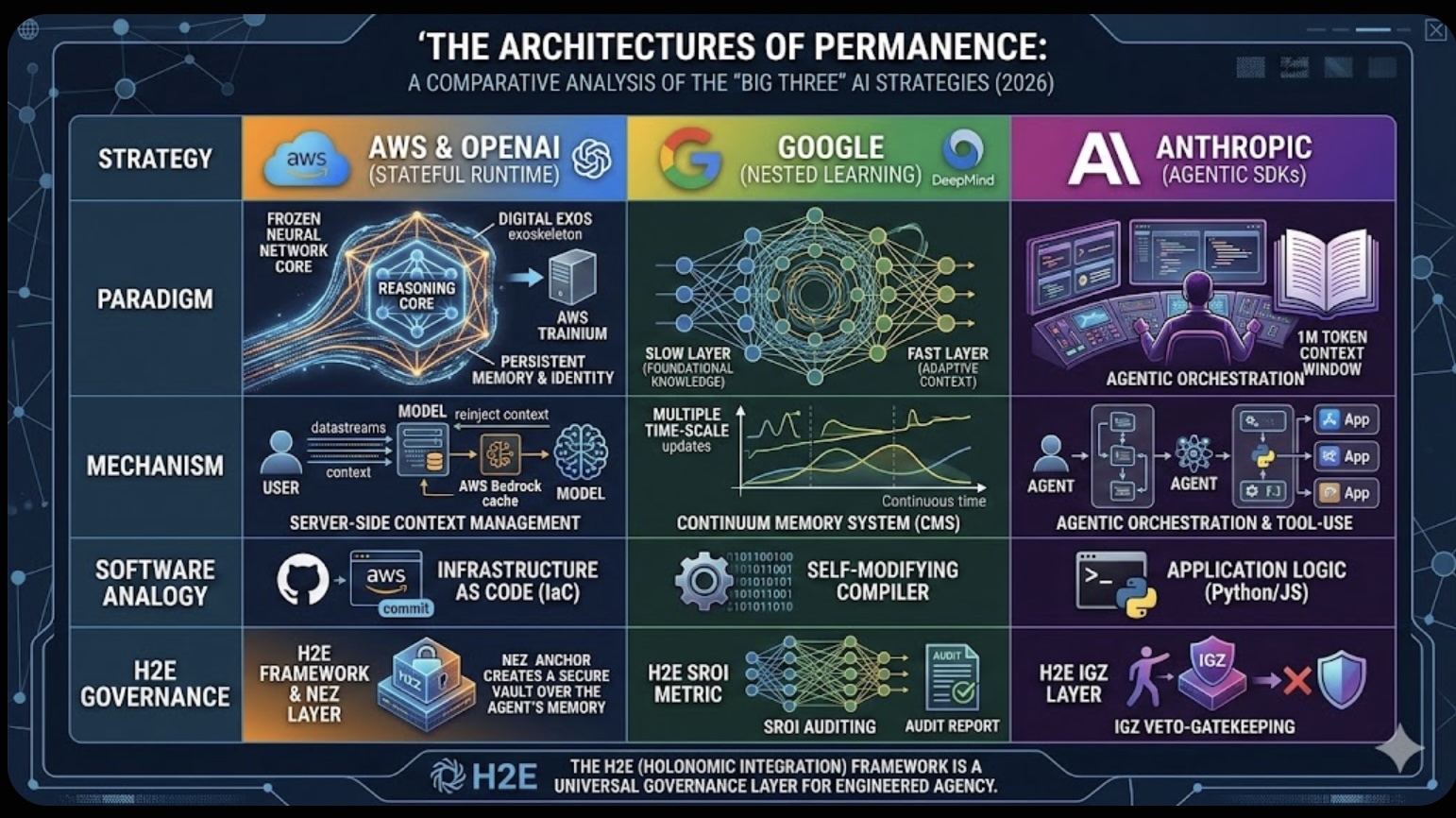 The Architectures of Permanence: A Comparative Analysis of the "Big Three" AI Strategies (2026)
The Architectures of Permanence: A Comparative Analysis of the "Big Three" AI Strategies (2026)